3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | Descriptor: | Divalent-cation tolerance protein cutA | | Authors: | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3AID
 
 | |
3APD
 
 | | Crystal structure of human PI3K-gamma in complex with CH5108134 | | Descriptor: | 5-(2-Morpholin-4-yl-7-pyridin-3-yl-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-pyrimidin-2-ylamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ATK
 
 | | Crystal structure of trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, DIMETHYL SULFOXIDE, ... | | Authors: | Yamane, J, Yao, M, Tanaka, I. | | Deposit date: | 2011-01-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3T03
 
 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
3AIK
 
 | | Crystal structure of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3AIN
 
 | | R267G mutant of a HSL-like carboxylesterase from Sulfolobus tokodaii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 303aa long hypothetical esterase, ... | | Authors: | Angkawidjaja, C, Kanaya, S. | | Deposit date: | 2010-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability of a thermostable carboxylesterase from the thermoacidophilic archaeon Sulfolobustokodaii
Febs J., 279, 2012
|
|
3D4Z
 
 | | GOLGI MANNOSIDASE II complex with gluco-imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, GLUCOIMIDAZOLE, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3B21
 
 | |
3B1M
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator Cerco-A | | Descriptor: | (9aS)-8-acetyl-N-[(2-ethylnaphthalen-1-yl)methyl]-1,7-dihydroxy-3-methoxy-9a-methyl-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Matsui, Y, Hiroyuki, H. | | Deposit date: | 2011-07-05 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pharmacology and in Vitro Profiling of a Novel Peroxisome Proliferator-Activated Receptor gamma Ligand, Cerco-A
Biol.Pharm.Bull., 34, 2011
|
|
3S1Y
 
 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3D51
 
 | | GOLGI MANNOSIDASE II complex with gluco-hydroxyiminolactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2Z21
 
 | |
3AA3
 
 | | A52L E. coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3AIL
 
 | |
3AA2
 
 | | A52I E. coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3AA5
 
 | | A52F E.coli RNase HI | | Descriptor: | Ribonuclease HI | | Authors: | Takano, K. | | Deposit date: | 2009-11-11 | | Release date: | 2010-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI
Protein Pept.Lett., 17, 2010
|
|
3D50
 
 | | GOLGI MANNOSIDASE II complex with N-octyl-6-epi-valienamine | | Descriptor: | (1S,2S,3R,6R)-4-(hydroxymethyl)-6-(octylamino)cyclohex-4-ene-1,2,3-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3QFQ
 
 | |
3D52
 
 | | GOLGI MANNOSIDASE II complex with an N-aryl carbamate derivative of gluco-hydroxyiminolactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2TEP
 
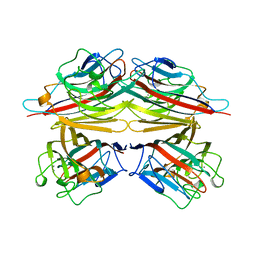 | | PEANUT LECTIN COMPLEXED WITH T-ANTIGENIC DISACCHARIDE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Ravindran, M, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-05 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Specificity of Peanut Agglutinin for Thomsen-Friedenreich Antigen is Mediated by Water-Bridges
Curr.Sci., 72, 1997
|
|
2WSF
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure determination and improved model of plant photosystem I.
J. Biol. Chem., 285, 2010
|
|
