4IDG
 
 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2
To be Published
|
|
1NAI
 
 | | UDP-GALACTOSE 4-EPIMERASE FROM ESCHERICHIA COLI, OXIDIZED | | Descriptor: | 1,3-PROPANDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Frey, P.A, Holden, H.M. | | Deposit date: | 1995-11-22 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli.
Biochemistry, 35, 1996
|
|
7ANH
 
 | | DdhaC | | Descriptor: | GDP-L-fucose synthase | | Authors: | Naismith, J.H, Woodward, L. | | Deposit date: | 2020-10-11 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | DdhaC
To Be Published
|
|
7ANC
 
 | |
8GIL
 
 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi (apo form) | | Descriptor: | L-threonine 3-dehydrogenase, POTASSIUM ION | | Authors: | Faria, J.N, Mercaldi, G.F, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
8GJB
 
 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi in complex with NAD and acetate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Mercaldi, G.F, Faria, J.N, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
7OL1
 
 | |
8H61
 
 | | Ketoreductase CpKR mutant - M2 | | Descriptor: | Mutant M2 of ketoreductase CpKR, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, C, Pan, J, Xu, J.H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational redesign of a robust ketoreductase for asymmetric synthesis of enantiopure diltiazem precursor.
To Be Published
|
|
6H0N
 
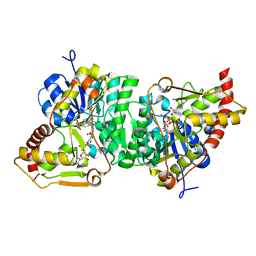 | |
6H0P
 
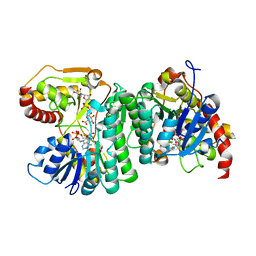 | |
3RFV
 
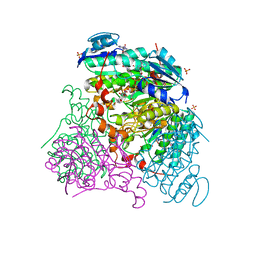 | | Crystal structure of Uronate dehydrogenase from Agrobacterium tumefaciens complexed with NADH and product | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-galactaro-1,5-lactone, PHOSPHATE ION, ... | | Authors: | Parkkinen, T, Rouvinen, J. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Uronate Dehydrogenase from Agrobacterium tumefaciens.
J.Biol.Chem., 286, 2011
|
|
3RFX
 
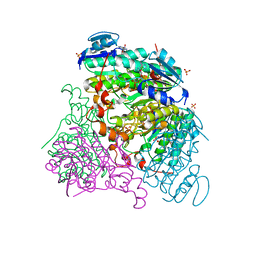 | |
3RFT
 
 | |
7KF3
 
 | |
7KN1
 
 | |
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
5GMO
 
 | | X-ray structure of carbonyl reductase SsCR | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Yu, H.L, Xu, J.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Efficient Synthesis of (R)-2-Chloro-1-(2,4-dichlorophenyl)ethanol with a Ketoreductase from Scheffersomyces stipitis CBS 6045
Adv.Synth.Catal., 359, 2017
|
|
7LL6
 
 | |
5GY7
 
 | | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis. | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NITRATE ION, ... | | Authors: | Singh, N, Tiwari, P, Phulera, S, Dixit, A, Choudhury, D. | | Deposit date: | 2016-09-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | X-Ray structure of H243I mutant of UDP-Galactose 4-epimerase from E.coli:evidence for existence of open and closed active site during catalysis.
To Be Published
|
|
3HFS
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | Descriptor: | Anthocyanidin reductase, CHLORIDE ION | | Authors: | Gargouri, M, Mauge, C, Langlois d'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ICP
 
 | |
3IUS
 
 | | The structure of a functionally unknown conserved protein from Silicibacter pomeroyi DSS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, uncharacterized conserved protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The structure of a functionally unknown conserved protein from Silicibacter pomeroyi DSS
To be Published
|
|
7M13
 
 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
1A9Z
 
 | |
1A9Y
 
 | |
