3DKP
 
 | | Human DEAD-box RNA-helicase DDX52, conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Wisniewska, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
3FHC
 
 | | Crystal structure of human Dbp5 in complex with Nup214 | | Descriptor: | ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | von Moeller, H, Conti, E. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner
Nat.Struct.Mol.Biol., 16, 2009
|
|
3FE2
 
 | | Human DEAD-BOX RNA helicase DDX5 (P68), conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX5, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases
Plos One, 5, 2010
|
|
3OIY
 
 | | Helicase domain of reverse gyrase from Thermotoga maritima | | Descriptor: | CHLORIDE ION, PYROPHOSPHATE 2-, reverse gyrase helicase domain | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-08-20 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The latch modulates nucleotide and DNA binding to the helicase-like domain of Thermotoga maritima reverse gyrase and is required for positive DNA supercoiling.
Nucleic Acids Res., 39, 2011
|
|
3MWL
 
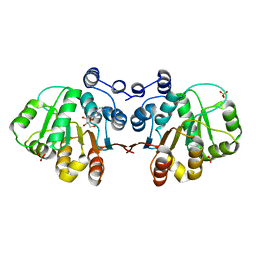 | | Q28E mutant of HERA N-terminal RecA-like domain in complex with 8-OXOADENOSINE | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-7H-purin-8-one, Heat resistant RNA dependent ATPase, SULFATE ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3MWJ
 
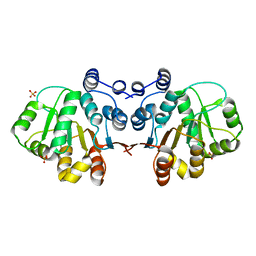 | |
3MWK
 
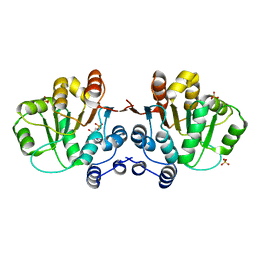 | | Q28E mutant of HERA N-terminal RecA-like domain, complex with 8-oxo-AMP | | Descriptor: | Heat resistant RNA dependent ATPase, SULFATE ION, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3NEJ
 
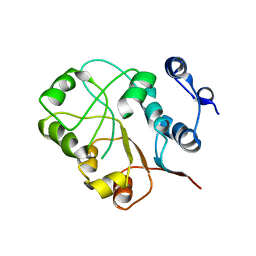 | |
7DTK
 
 | |
5GJU
 
 | | DEAD-box RNA helicase | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Li, F, Wang, L, Shi, Y. | | Deposit date: | 2016-07-02 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5GVS
 
 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
5GVR
 
 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
5H1Y
 
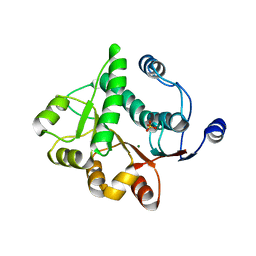 | |
5ZBZ
 
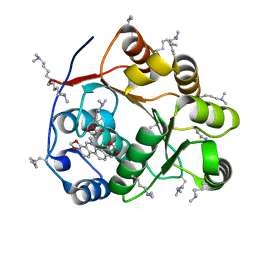 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
6AIB
 
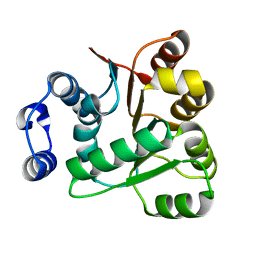 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6AIC
 
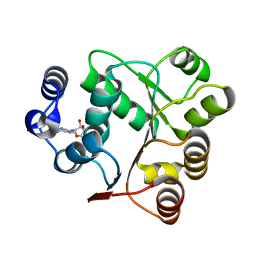 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8PNK
 
 | |
4W7S
 
 | | Crystal structure of the yeast DEAD-box splicing factor Prp28 at 2.54 Angstroms resolution | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jacewicz, A, Smith, P, Schwer, B, Shuman, S. | | Deposit date: | 2014-08-22 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Crystal structure, mutational analysis and RNA-dependent ATPase activity of the yeast DEAD-box pre-mRNA splicing factor Prp28.
Nucleic Acids Res., 42, 2014
|
|
4F93
 
 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4XGT
 
 | |
1S2M
 
 | |
4ON9
 
 | | DECH box helicase domain | | Descriptor: | CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58, SULFATE ION | | Authors: | Deimling, T, Witte, G, Hopfner, K.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal and solution structure of the human RIG-I SF2 domain
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7SSG
 
 | | Mfd DNA complex | | Descriptor: | DNA (5'-D(P*TP*GP*GP*CP*GP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*GP*CP*CP*TP*CP*GP*CP*TP*GP*CP*CP*A)-3'), Transcription-repair-coupling factor | | Authors: | Oakley, A.J, Xu, Z.-Q. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Mechanism of transcription modulation by the transcription-repair coupling factor.
Nucleic Acids Res., 50, 2022
|
|
5LBA
 
 | | Crystal structure of human RECQL5 helicase in complex with DSPL fragment(1-cyclohexyl-3-(oxolan-2-ylmethyl)urea, SGC - Diamond XChem I04-1 fragment screening. | | Descriptor: | 1-cyclohexyl-3-[[(2~{R})-oxolan-2-yl]methyl]urea, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, ... | | Authors: | Newman, J.A, Aitkenhead, H, Talon, R, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human RECQL5 helicase in complex with 3D fragment (1-cyclohexyl-3-(oxolan-2-ylmethyl)urea)
To be published
|
|
4KBG
 
 | |
