8X6P
 
 | | Isomerase Protein | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Guven, O, Demirci, H. | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Isomerase Protein
To Be Published
|
|
5J6E
 
 | |
5L1D
 
 | |
3CGN
 
 | | Crystal Structure of thermophilic SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Neumann, P, Loew, C, Stubbs, M.T, Balbach, J. | | Deposit date: | 2008-03-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
3EY6
 
 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
7OXI
 
 | | ttSlyD with W4A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXK
 
 | | ttSlyD with W4K pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXG
 
 | | ttSlyD FKBP domain with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXJ
 
 | | ttSlyD with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Fragment of 30S ribosomal protein S2 peptide, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXH
 
 | | ttSlyD with pseudo-wild-type S2 peptide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 30S ribosomal protein S2, CHLORIDE ION, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
3JYM
 
 | |
3UQI
 
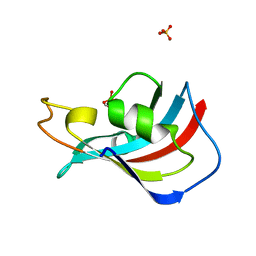 | |
3JXV
 
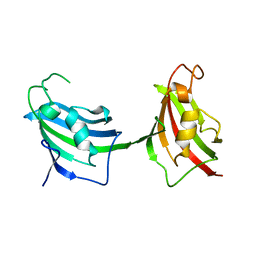 | |
2VN1
 
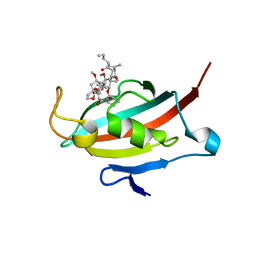 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | Descriptor: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
6TX7
 
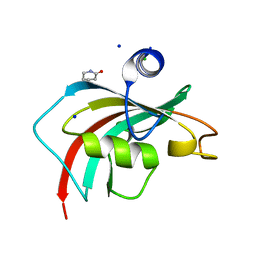 | |
6TX8
 
 | |
6TX6
 
 | |
6TX5
 
 | |
3PA7
 
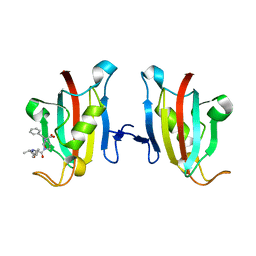 | | Crystal structure of FKBP from plasmodium vivax in complex with tetrapeptide ALPF | | Descriptor: | 4-mer Peptide ALPF, 70 kDa peptidylprolyl isomerase, putative | | Authors: | Balakrishna, A.M, Alag, R, Yoon, H.S. | | Deposit date: | 2010-10-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax
EUKARYOTIC CELL, 12, 2013
|
|
6TXX
 
 | | CRYSTAL STRUCTURE OF HUMAN FKBP51 FK1 DOMAIN A19T MUTANT IN COMPLEX WITH SAFit2 | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Fiegen, D, Draxler, S.W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Hybrid Screening Approach for Very Small Fragments: X-ray and Computational Screening on FKBP51.
J.Med.Chem., 63, 2020
|
|
7JMF
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 42 - State 6 (S6) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
6SAF
 
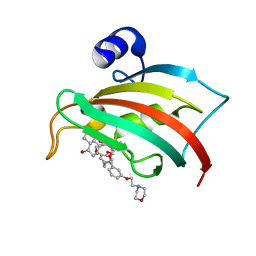 | | The Fk1 domain of FKBP51 in complex with (S)-(R)-3-(3,4-dimethoxyphenyl)-1-(3-(2-morpholinoethoxy)phenyl)propyl 1-((1R,4aR,8aR)-4-oxodecahydronaphthalene-1-carbonyl)piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [(1~{R})-3-(3,4-dimethoxyphenyl)-1-[3-(2-morpholin-4-ylethoxy)phenyl]propyl] (2~{S})-1-[[(1~{R},4~{a}~{R},8~{a}~{R})-4-oxidanylidene-2,3,4~{a},5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-1-yl]carbonyl]piperidine-2-carboxylate | | Authors: | Feng, X, Sippel, C, Knaup, F, Bracher, A, Staibano, S, Romano, M.F, Hausch, F. | | Deposit date: | 2019-07-16 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Novel Decalin-Based Bicyclic Scaffold for FKBP51-Selective Ligands.
J.Med.Chem., 63, 2020
|
|
6MKE
 
 | |
5DIT
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
