6VPL
 
 | |
6UVM
 
 | | Cocrystal of BRD4(D1) with a methyl carbamate thiazepane inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[(7S)-7-(thiophen-2-yl)-6,7-dihydro-1,4-thiazepin-4(5H)-yl]ethan-1-one, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Pomerantz, W.C.K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluating the Advantages of Using 3D-Enriched Fragments for Targeting BET Bromodomains.
Acs Med.Chem.Lett., 10, 2019
|
|
6V17
 
 | | Crystal structure of the bromodomain of human BRD7 bound to I-BRD9 | | Descriptor: | Bromodomain-containing protein 7, CHLORIDE ION, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V0U
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1F
 
 | | Crystal structure of the bromodomain of human BRD7 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6W1S
 
 | | Atomic model of the mammalian Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Young, N, Asturias, F. | | Deposit date: | 2020-03-04 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
Cell, 178, 2019
|
|
6TPX
 
 | |
6VCS
 
 | | SRA domain of UHRF1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*AP*CP*AP*GP*GP*C)-3'), E3 ubiquitin-protein ligase UHRF1, UNK-UNK-UNK-UNK, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SRA domain of UHRF1 in complex with DNA
To Be Published
|
|
6WBW
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6WG3
 
 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6X3Y
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-21 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6X4N
 
 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 24) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-3-oxa-9,12,15,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
6WBZ
 
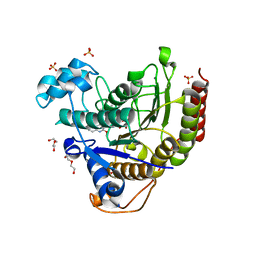 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | Descriptor: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X4O
 
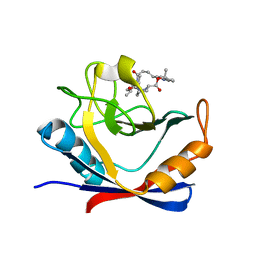 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,21,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(26),18,20,22,24,27-hexaene-4,10,13,16-tetrone (compound 21) | | Descriptor: | (2R,5S,11S,14S,18E)-2,11-dimethyl-14-(propan-2-yl)-3-oxa-9,12,15,21,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(26),18,20,22,24,27-hexaene-4,10,13,16-tetrone, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
5ORV
 
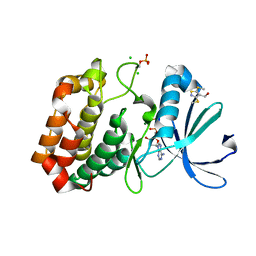 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OS3
 
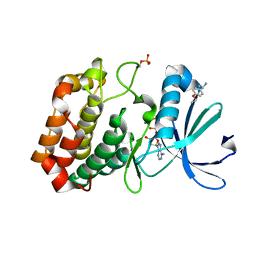 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | (1~{R})-1-(4-ethoxyphenyl)ethanamine, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OS4
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | (3~{a}~{R},5~{S},7~{a}~{S})-5-phenyl-3~{a},4,5,6,7,7~{a}-hexahydroisoindole-1,3-dione, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5ORL
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, CHLORIDE ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, Vrzal, L, Birchall, K, Arnold, L.H, Mpamhanga, C, Coombs, P.J, Burgess, S.G, Richards, M.W, Winter, A, Veverka, V, von Delft, F, Merritt, A, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OWW
 
 | | Crystal structure of human BRD4(1) bromodomain in complex with UT22B | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-methylbenzotriazol-5-yl)-1-(phenylmethyl)imidazole-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-09-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
5ORY
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 2,4-bis(fluoranyl)-6-(1~{H}-pyrazol-3-yl)phenol, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OSF
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 2-(4-ethylphenoxy)-1-piperidin-1-yl-ethanone, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-17 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5ORS
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5ORW
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 3-(4-fluoranylphenoxy)-1-thiomorpholin-4-yl-propan-1-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OS5
 
 | | Crystal structure of Aurora-A kinase in complex with an allosterically binding fragment | | Descriptor: | 4-(4-hydroxyphenyl)sulfanylphenol, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | McIntyre, P.J, Collins, P.M, von Delft, F, Bayliss, R. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-01 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Characterization of Three Druggable Hot-Spots in the Aurora-A/TPX2 Interaction Using Biochemical, Biophysical, and Fragment-Based Approaches.
ACS Chem. Biol., 12, 2017
|
|
5OV3
 
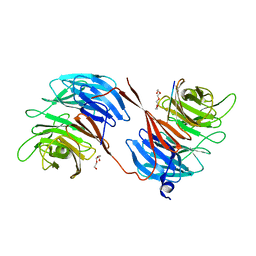 | | Structure of the RbBP5 beta-propeller domain | | Descriptor: | Retinoblastoma-binding protein 5, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Mittal, A, Zhang, Y, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2017-08-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the RbBP5 beta-propeller domain reveals a surface with potential nucleic acid binding sites.
Nucleic Acids Res., 46, 2018
|
|
