1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
4AKJ
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4P5T
 
 | | 14.C6 TCR complexed with MHC class II I-Ab/3K peptide | | Descriptor: | H-2 class II histocompatibility antigen, A-B alpha chain, Human nkt tcr beta chain, ... | | Authors: | Trenh, P, Stadinski, B, Huseby, E.S, Stern, L.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
2XQY
 
 | | CRYSTAL STRUCTURE OF PSEUDORABIES CORE FRAGMENT OF GLYCOPROTEIN H IN COMPLEX WITH FAB D6.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A13-D6.3 MONOCLONAL ANTIBODY, ... | | Authors: | Backovic, M, Dubois, R, Cockburn, J, Sharff, A, Vaney, M, Granzow, H, Klupp, B, Bricogne, G, Mettenleiter, T, Rey, F. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a Core Fragment of Glycoprotein H from Pseudorabies Virus in Complex with Antibody.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5BTI
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1UY4
 
 | |
1UY3
 
 | |
1NHT
 
 | | ENTRAPMENT OF 6-THIOPHOSPHORYL-IMP IN THE ACTIVE SITE OF CRYSTALLINE ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI DATA COLLECTED AT 100K | | Descriptor: | 2-DEAZO-6-THIOPHOSPHATE GUANOSINE-5'-MONOPHOSPHATE, ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Poland, B.W, Bruns, C.A, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 1997-01-12 | | Release date: | 1997-10-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Entrapment of 6-thiophosphoryl-IMP in the active site of crystalline adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 272, 1997
|
|
5CTN
 
 | | Structure of BPu1 beta-lactamase | | Descriptor: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-25 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
7X1B
 
 | |
7X1C
 
 | |
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
3PBY
 
 | | Crystal structure of the mutant L123S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBW
 
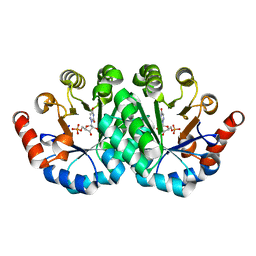 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBU
 
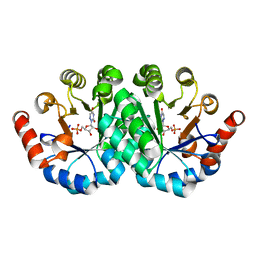 | | Crystal structure of the mutant I96S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
5DKP
 
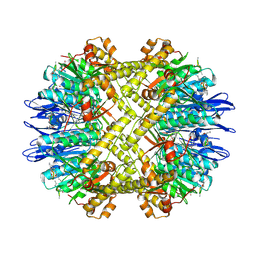 | | Crystal Structure of N. meningitidis ClpP in complex with agonist ADEP A54556. | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION, SODIUM ION, ... | | Authors: | Goodreid, J.D, Janetzko, J, Santa Maria Jr, J.P, Wong, K, Leung, E, Eger, B.T, Bryson, S, Pai, E.F, Gray-Owen, S.D, Walker, S, Houry, W.A, Batey, R.A. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Development and Characterization of Potent Cyclic Acyldepsipeptide Analogues with Increased Antimicrobial Activity.
J.Med.Chem., 59, 2016
|
|
5KLC
 
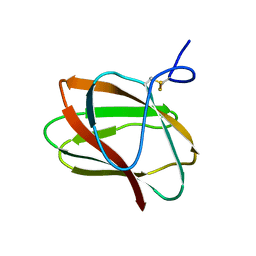 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome | | Descriptor: | Carbohydrate binding module E1 | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
3PBV
 
 | | Crystal structure of the mutant I96T of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3KW7
 
 | | Crystal structure of LacB from Trametes sp. AH28-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ge, H.H, Teng, M.K, Niu, L.W. | | Deposit date: | 2009-12-01 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure of native laccase B from Trametes sp. AH28-2
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7Y79
 
 | | Crystal structure of Cry78Aa | | Descriptor: | Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
6I5A
 
 | | Tobacco Mosaic Virus | | Descriptor: | Capsid protein | | Authors: | Song, B, Flegler, V, Makbul, C, Rasmussen, T, Bottcher, B. | | Deposit date: | 2018-11-13 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Capabilities of the Falcon III detector for single-particle structure determination.
Ultramicroscopy, 203, 2019
|
|
6H6R
 
 | | Fragment Derived XIAP inhibitor | | Descriptor: | 2-[[(2~{R},5~{R})-1-[2-[6-[(4-fluorophenyl)methyl]-3,3-dimethyl-2~{H}-pyrrolo[3,2-b]pyridin-1-yl]-2-oxidanylidene-ethyl]-5-methyl-piperazin-2-yl]methyl]-3~{H}-isoindol-1-one, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Williams, P.A. | | Deposit date: | 2018-07-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Fragment-Derived Clinical Candidate for Antagonism of X-Linked and Cellular Inhibitor of Apoptosis Proteins: 1-(6-[(4-Fluorophenyl)methyl]-5-(hydroxymethyl)-3,3-dimethyl-1 H,2 H,3 H-pyrrolo[3,2- b]pyridin-1-yl)-2-[(2 R,5 R)-5-methyl-2-([(3R)-3-methylmorpholin-4-yl]methyl)piperazin-1-yl]ethan-1-one (ASTX660).
J. Med. Chem., 61, 2018
|
|
2IBZ
 
 | | Yeast Cytochrome BC1 Complex with Stigmatellin | | Descriptor: | 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, Cytochrome b, Cytochrome c1, ... | | Authors: | Hunte, C. | | Deposit date: | 2006-09-12 | | Release date: | 2007-03-20 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
3I54
 
 | | Crystal structure of MtbCRP in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, Crp/Fnr family | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
5POV
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11063a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-methylthieno[2,3-b]pyridine-2-carboxamide, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
