7YRK
 
 | | Crystal structure of the hen egg lysozyme-2-oxidobenzylidene-threoninato-copper (II) complex | | Descriptor: | (6~{S})-6-[(1~{R})-1-oxidanylethyl]-2,4-dioxa-7$l^{4}-aza-3$l^{3}-cupratricyclo[7.4.0.0^{3,7}]trideca-1(13),7,9,11-tetraen-5-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Unno, M, Furuya, T, Kitanishi, K, Akitsu, T. | | Deposit date: | 2022-08-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A novel hybrid protein composed of superoxide-dismutase-active Cu(II) complex and lysozyme.
Sci Rep, 13, 2023
|
|
2PPN
 
 | |
6ETN
 
 | |
3X0J
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in apo state at 0.92 angstrom resolution | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, SULFATE ION | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-16 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
6F7R
 
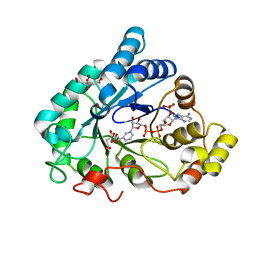 | | AKR1B1 at 0.03 MGy radiation dose. | | Descriptor: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Castellvi, A, Juanhuix, J. | | Deposit date: | 2017-12-11 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
4E1U
 
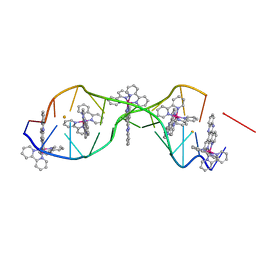 | | [Ru(bpy)2 dppz]2+ bound to DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*AP*CP*CP*G)-3', BARIUM ION, Delta-[Ru(bpy)2dppz]2+ | | Authors: | Song, H, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Crystal structure of delta-[Ru(bpy)2dppz]2+ bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation.
Nat Chem, 4, 2012
|
|
2XJH
 
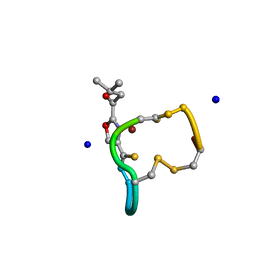 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
3GYJ
 
 | |
7A2X
 
 | |
2QDW
 
 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
1PWM
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
5DKM
 
 | |
4RJ1
 
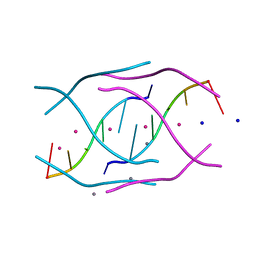 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
6UWW
 
 | |
4AR6
 
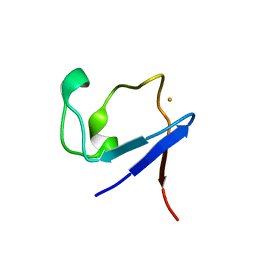 | | X-ray crystallographic structure of the reduced form perdeuterated Pyrococcus furiosus rubredoxin at 295 K (in quartz capillary) to 0.92 Angstroms resolution. | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Cuypers, M.G, Mason, S.A, Blakeley, M.P, Mitchell, E.P, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2012-04-20 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Near-Atomic Resolution Neutron Crystallography on Perdeuterated Pyrococcus Furiosus Rubredoxin: Implication of Hydronium Ions and Protonation Equilibria and Hydronium Ions in Redox Changes
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5RCK
 
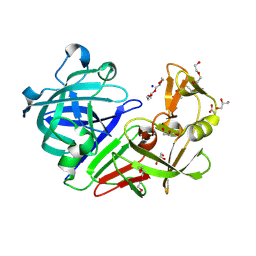 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 05 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
2G6F
 
 | |
5R33
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 27, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1L9L
 
 | | GRANULYSIN FROM HUMAN CYTOLYTIC T LYMPHOCYTES | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ETHANOL, Granulysin, ... | | Authors: | Anderson, D.H, Sawaya, M.R, Cascio, D, Ernst, W, Krensky, A, Modlin, R, Eisenberg, D. | | Deposit date: | 2002-03-25 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Granulysin Crystal Structure and a Structure-Derived Lytic Mechanism
J.Mol.Biol., 325, 2002
|
|
7XBC
 
 | | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lignoceric acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | The 0.92 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lignoceric acid
To Be Published
|
|
6CNW
 
 | | STRUCTURE OF HUMANIZED SINGLE DOMAIN ANTIBODY SD84 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, humanized antibody SD84h | | Authors: | Luo, J, Obmolova, O. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
2GBA
 
 | | Reduced Cu(I) form at pH 4 of P52G mutant of amicyanin | | Descriptor: | COPPER (I) ION, amicyanin | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
1IQZ
 
 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM I) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
8PB7
 
 | | PsiM in complex with sinefungin and baeocystin | | Descriptor: | Baeocystin, CHLORIDE ION, Psilocybin synthase, ... | | Authors: | Werten, S, Hudspeth, J, Rupp, B. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Methyl transfer in psilocybin biosynthesis.
Nat Commun, 15, 2024
|
|
2FVY
 
 | | High Resolution Glucose Bound Crystal Structure of GGBP | | Descriptor: | ACETATE ION, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2006-01-31 | | Release date: | 2007-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures.
Protein Sci., 16, 2007
|
|
