5KKI
 
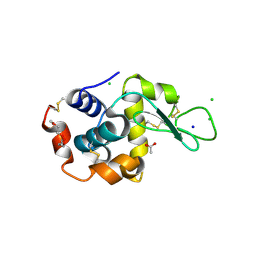 | | 1.7-Angstrom in situ Mylar structure of hen egg-white lysozyme (HEWL) at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Broecker, J, Ernst, O.P. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Versatile System for High-Throughput In Situ X-ray Screening and Data Collection of Soluble and Membrane-Protein Crystals.
Cryst Growth Des, 16, 2016
|
|
8JO4
 
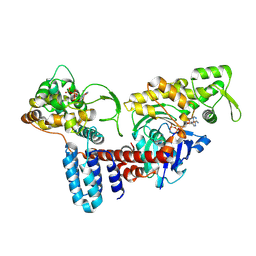 | | Cryo-EM structure of a Legionella effector complexed with actin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8JO3
 
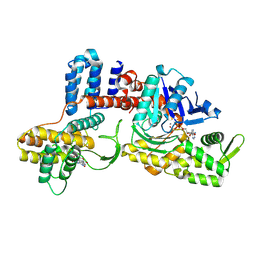 | | Cryo-EM structure of a Legionella effector complexed with actin and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
7MX3
 
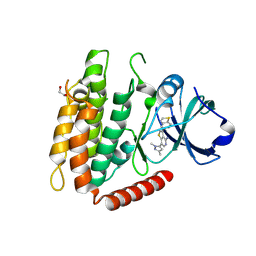 | | Crystal structure of human RIPK3 complexed with GSK'843 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
7UCS
 
 | |
6B80
 
 | | Crystal structure of myotoxin II from Bothrops moojeni complexed to myristic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog 2, MYRISTIC ACID, ... | | Authors: | Salvador, G.H.M, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural evidence for a fatty acid-independent myotoxic mechanism for a phospholipase A2-like toxin.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
7UCT
 
 | |
7UD3
 
 | |
7UCV
 
 | |
5KPK
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
6BD5
 
 | | Crystal structure of human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl (2-{[(2R)-2-(cyclopentylamino)-3-oxo-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}ethyl)carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2017-10-21 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of Human CYP3A4 by Rationally Designed Ritonavir-Like Compounds: Impact and Interplay of the Side Group Functionalities.
Mol Pharm., 15, 2018
|
|
7UCZ
 
 | |
7UD1
 
 | |
8EGI
 
 | | X-ray structure of carbonmonoxy hemoglobin in complex with VZHE039-NO | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-09-12 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Investigation of Novel Nitric Oxide (NO)-Releasing Aromatic Aldehydes as Drug Candidates for the Treatment of Sickle Cell Disease.
Molecules, 27, 2022
|
|
7UCN
 
 | |
5KQY
 
 | | Protease E35D-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease E35D-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
8JY0
 
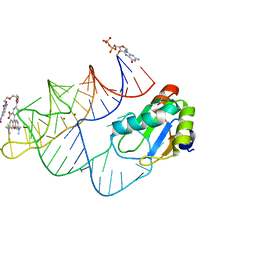 | | Crystal structure of RhoBAST complexed with TMR-DN | | Descriptor: | 2,4-dinitroaniline, 5-aminocarbonyl-2-[3-(dimethylamino)-6-dimethylazaniumylidene-xanthen-9-yl]benzoate, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Xiao, Y, Xu, Z, Fang, X. | | Deposit date: | 2023-07-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms for binding and activation of a contact-quenched fluorophore by RhoBAST.
Nat Commun, 15, 2024
|
|
6BFN
 
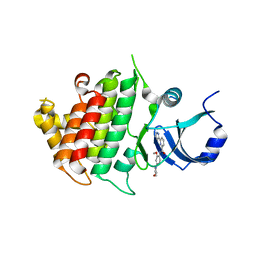 | | Crystal structure of human IRAK1 | | Descriptor: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | Authors: | Wang, L, Qiao, Q, Wu, H. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6B1Q
 
 | |
6B36
 
 | |
5KTH
 
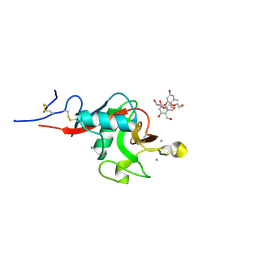 | | Structure of cow mincle complexed with brartemicin | | Descriptor: | 2,4-dihydroxy-6-methyl Benzoic acid, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Feinberg, H, Rambaruth, N.D.S, Jegouzo, S.A.F, Jacobsen, K.M, Djurhuus, R, Poulsen, T.B, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
5L2Q
 
 | |
5L3H
 
 | |
6B3C
 
 | |
5KU8
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D, Dowling, J. | | Deposit date: | 2016-07-13 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of CK2
Not Published
|
|
