1YFG
 
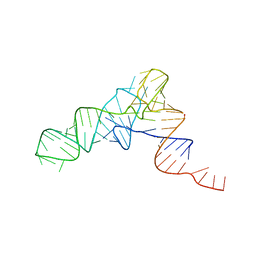 | | YEAST INITIATOR TRNA | | Descriptor: | YEAST INITIATOR TRNA | | Authors: | Basavappa, R, Sigler, P.B. | | Deposit date: | 1997-05-08 | | Release date: | 1997-09-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination.
EMBO J., 10, 1991
|
|
2AXD
 
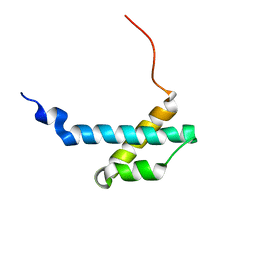 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
1P64
 
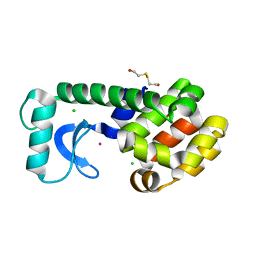 | | T4 LYSOZYME CORE REPACKING MUTANT L133F/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-28 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1YI5
 
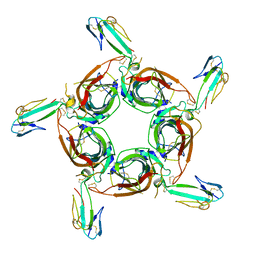 | | Crystal structure of the a-cobratoxin-AChBP complex | | Descriptor: | Acetylcholine-binding protein, Long neurotoxin 1 | | Authors: | Bourne, Y, Talley, T.T, Hansen, S.B, Taylor, P, Marchot, P. | | Deposit date: | 2005-01-11 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors
Embo J., 24, 2005
|
|
2B75
 
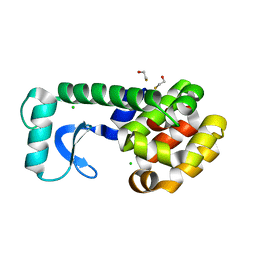 | | T4 Lysozyme mutant L99A at 150 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1P6M
 
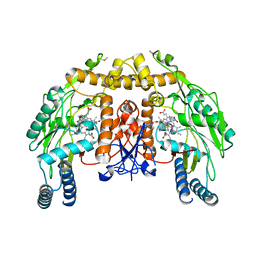 | | Bovine endothelial NOS heme domain with (4S)-N-(4-amino-5-[aminoethyl]aminopentyl)-N'-nitroguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2B70
 
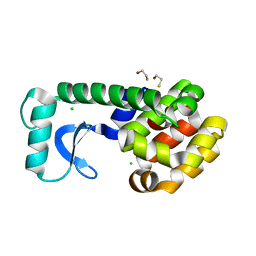 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6Z
 
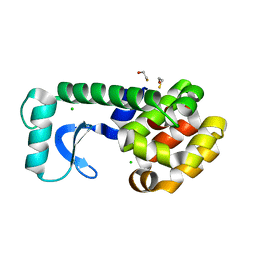 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2BBP
 
 | |
1Y3I
 
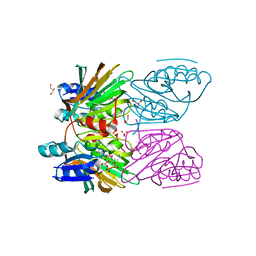 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | Descriptor: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
2B4G
 
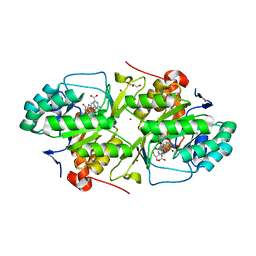 | | dihydroorotate dehydrogenase | | Descriptor: | BROMIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Arakaki, T.L, Merritt, E.A, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies
Mol.Microbiol., 68, 2008
|
|
2AZ3
 
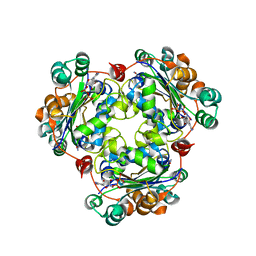 | | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum in complex with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Besir, H, Zeth, K, Bracher, A, Heider, U, Ishibashi, M, Tokunaga, M, Oesterhelt, D. | | Deposit date: | 2005-09-09 | | Release date: | 2005-12-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a halophilic nucleoside diphosphate kinase from Halobacterium salinarum
Febs Lett., 579, 2005
|
|
2B0G
 
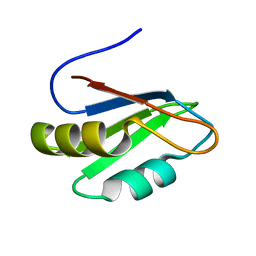 | |
2B5D
 
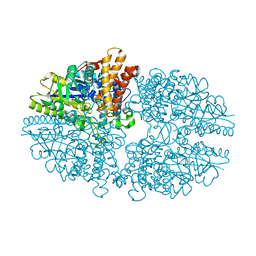 | | Crystal structure of the novel alpha-amylase AmyC from Thermotoga maritima | | Descriptor: | alpha-Amylase | | Authors: | Dickmanns, A, Ballschmiter, M, Liebl, W, Ficner, R. | | Deposit date: | 2005-09-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the novel alpha-amylase AmyC from Thermotoga maritima.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1ORS
 
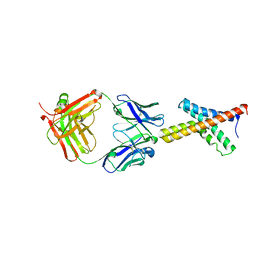 | | X-ray structure of the KvAP potassium channel voltage sensor in complex with an Fab | | Descriptor: | 33H1 Fab heavy chain, 33H1 Fab light chain, potassium channel | | Authors: | Jiang, Y, Lee, A, Chen, J, Ruta, V, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2003-03-14 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of a voltage-dependent K+ channel
Nature, 423, 2003
|
|
1YJM
 
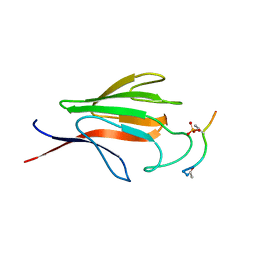 | | Crystal structure of the FHA domain of mouse polynucleotide kinase in complex with an XRCC4-derived phosphopeptide. | | Descriptor: | 12-mer peptide from DNA-repair protein XRCC4, Polynucleotide 5'-hydroxyl-kinase | | Authors: | Bernstein, N.K, Williams, R.S, Rakovszky, M.L, Cui, D, Green, R, Karimi-Busheri, F, Mani, R.S, Galicia, S, Koch, C.A, Cass, C.E, Durocher, D, Weinfeld, M, Glover, J.N.M. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase.
Mol.Cell, 17, 2005
|
|
1OVE
 
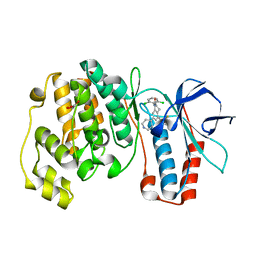 | | The structure of p38 alpha in complex with a dihydroquinolinone | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERIDIN-4-YL-3,4-DIHYDROQUINOLIN-2(1H)-ONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
2AA9
 
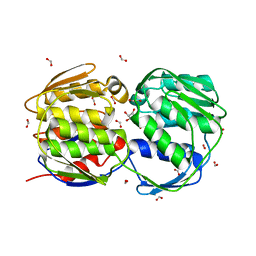 | | EPSP synthase liganded with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID | | Authors: | Priestman, M.A, Healy, M.L, Funke, T, Becker, A, Schonbrunn, E. | | Deposit date: | 2005-07-13 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate.
Febs Lett., 579, 2005
|
|
2A7K
 
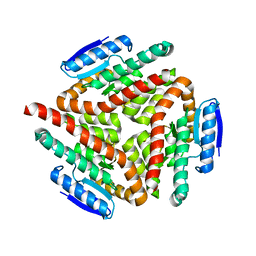 | | carboxymethylproline synthase (CarB) from pectobacterium carotovora, apo enzyme | | Descriptor: | CarB | | Authors: | Sleeman, M.C, Sorensen, J.L, Batchelar, E.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Mechanistic Studies on Carboxymethylproline Synthase (CarB), a Unique Member of the Crotonase Superfamily Catalyzing the First Step in Carbapenem Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
1OY7
 
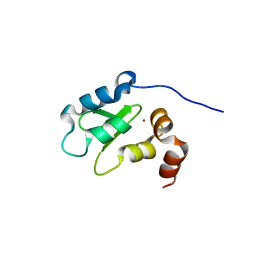 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEVVAVKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1P6N
 
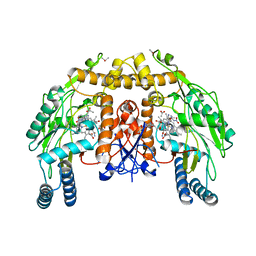 | | Bovine endothelial NOS heme domain with L-N(omega)-nitroarginine-(4R)-amino-L-proline amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1P1L
 
 | |
1YG0
 
 | |
1P5D
 
 | |
232D
 
 | |
