1DPQ
 
 | |
1DPR
 
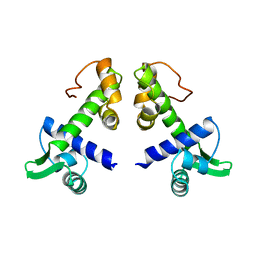 | | STRUCTURES OF THE APO-AND METAL ION ACTIVATED FORMS OF THE DIPHTHERIA TOX REPRESSOR FROM CORYNEBACTERIUM DIPHTHERIAE | | Descriptor: | DIPHTHERIA TOX REPRESSOR | | Authors: | Schiering, N, Tao, X, Murphy, J, Petsko, G.A, Ringe, D. | | Deposit date: | 1995-02-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1DPS
 
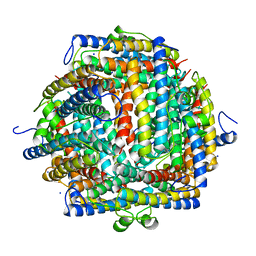 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1DPT
 
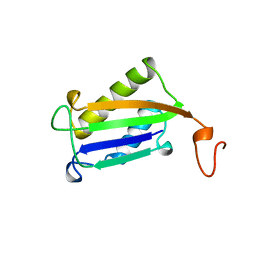 | | D-DOPACHROME TAUTOMERASE | | Descriptor: | D-DOPACHROME TAUTOMERASE | | Authors: | Sugimoto, H, Taniguchi, M, Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-05-11 | | Release date: | 1999-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of human D-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 A resolution.
Biochemistry, 38, 1999
|
|
1DPU
 
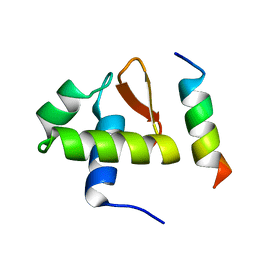 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN RPA32 COMPLEXED WITH UNG2(73-88) | | Descriptor: | REPLICATION PROTEIN A (RPA32) C-TERMINAL DOMAIN, URACIL DNA GLYCOSYLASE (UNG2) | | Authors: | Mer, G, Edwards, A.M, Chazin, W.J. | | Deposit date: | 1999-12-27 | | Release date: | 2000-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA.
Cell(Cambridge,Mass.), 103, 2000
|
|
1DPW
 
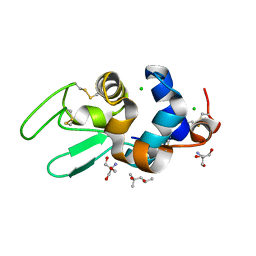 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME IN COMPLEX WITH MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DPX
 
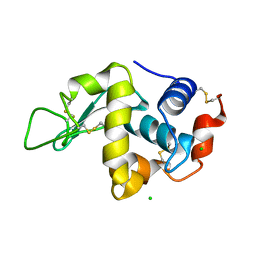 | | STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Weiss, M.S, Palm, G.J, Hilgenfeld, R. | | Deposit date: | 1999-12-28 | | Release date: | 2000-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization, structure solution and refinement of hen egg-white lysozyme at pH 8.0 in the presence of MPD.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DPY
 
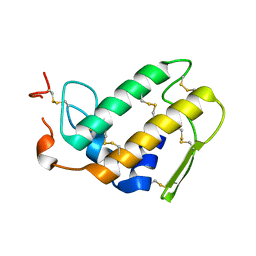 | | THREE-DIMENSIONAL STRUCTURE OF A NOVEL PHOSPHOLIPASE A2 FROM INDIAN COMMON KRAIT AT 2.45 A RESOLUTION | | Descriptor: | PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Singh, G, Gourinath, S, Sharma, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-28 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Sequence and crystal structure determination of a basic phospholipase A2 from common krait (Bungarus caeruleus) at 2.4 A resolution: identification and characterization of its pharmacological sites.
J.Mol.Biol., 307, 2001
|
|
1DPZ
 
 | | STRUCTURE OF MODIFIED 3-ISOPROPYLMALATE DEHYDROGENASE AT THE C-TERMINUS, HD711 | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Nurachman, Z, Akanuma, S, Sato, T, Oshima, T, Tanaka, N. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 3-isopropylmalate dehydrogenases with mutations at the C-terminus: crystallographic analyses of structure-stability relationships.
Protein Eng., 13, 2000
|
|
1DQ0
 
 | | Locked, metal-free concanavalin A, a minor species in solution | | Descriptor: | Concanavalin-Br | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ1
 
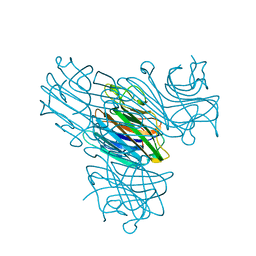 | | Calcium;Calcium concanavalin A | | Descriptor: | CALCIUM ION, Concanavalin-Br | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ2
 
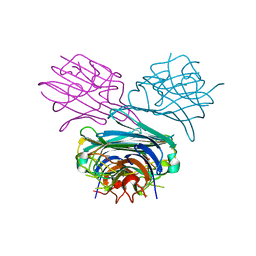 | | Unlocked metal-free concanavalin A | | Descriptor: | ACETIC ACID, Concanavalin-Br | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ3
 
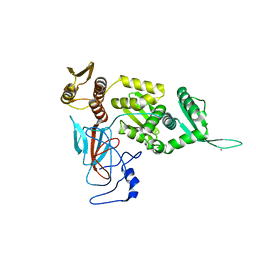 | |
1DQ4
 
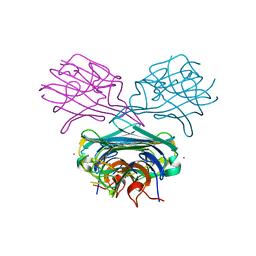 | | A transient unlocked concanavalin A structure with MN2+ bound in the transition metal ion binding site S1 and an empty calcium binding site S2 | | Descriptor: | Concanavalin-Br, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ5
 
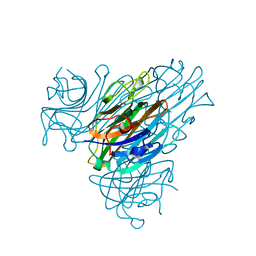 | | Manganese;Manganese concanavalin A at pH 5.0 | | Descriptor: | Concanavalin-Br, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ6
 
 | | Manganese;Manganese concanavalin A at pH 7.0 | | Descriptor: | Concanavalin-A, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
1DQ7
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NEUROTOXIN FROM RED SCORPION (BUTHUS TAMULUS) AT 2.2A RESOLUTION. | | Descriptor: | NEUROTOXIN | | Authors: | Sharma, M, Yadav, S, Karthikeyan, S, Kumar, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-30 | | Release date: | 2000-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional Structure of a Neurotoxin from Red Scorpion (Buthus tamulus) at 2.2A Resolution
To be Published
|
|
1DQ8
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG AND COA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQ9
 
 | | COMPLEX OF CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG-COA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, PROTEIN (HMG-COA REDUCTASE) | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQA
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG, COA, AND NADP+ | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1DQB
 
 | |
1DQC
 
 | | SOLUTION STRUCTURE OF TACHYCITIN, AN ANTIMICROBIAL PROTEIN WITH CHITIN-BINDING FUNCTION | | Descriptor: | TACHYCITIN | | Authors: | Suetake, T, Tsuda, S, Kawabata, S, Miura, K, Kawano, K. | | Deposit date: | 2000-01-04 | | Release date: | 2000-09-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif.
J.Biol.Chem., 275, 2000
|
|
1DQD
 
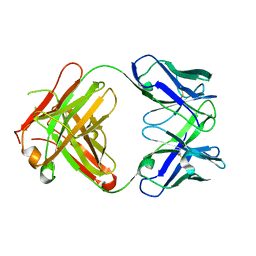 | | CRYSTAL STRUCTURE OF FAB HGR-2 F6, A COMPETITIVE ANTAGONIST OF THE GLUCAGON RECEPTOR | | Descriptor: | FAB HGR-2 F6 | | Authors: | Wright, L.M, Brzozowski, A.M, Hubbard, R.E, Pike, A.C.W, Roberts, S.M, Skovgaard, R.N, Svendsen, I, Vissing, H, Bywater, R.P. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Fab hGR-2 F6, a competitive antagonist of the glucagon receptor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DQE
 
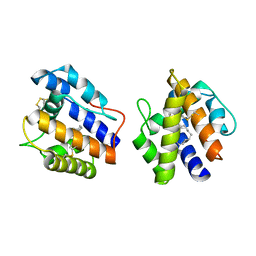 | | BOMBYX MORI PHEROMONE BINDING PROTEIN | | Descriptor: | HEXADECA-10,12-DIEN-1-OL, PHEROMONE-BINDING PROTEIN | | Authors: | Sandler, B.H, Nikonova, L, Leal, W.S, Clardy, J. | | Deposit date: | 2000-01-04 | | Release date: | 2001-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex.
Chem.Biol., 7, 2000
|
|
1DQF
 
 | |
