6EFR
 
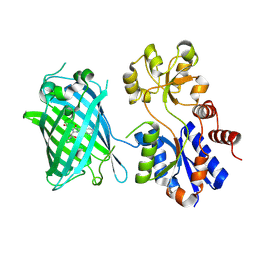 | | Crystal Structure of iNicSnFR 1.0 | | Descriptor: | iNicSnFR 1.0, a genetically encoded nicotine biosensor,Green fluorescent protein | | Authors: | Shivange, A.V, Borden, P.M. | | Deposit date: | 2018-08-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nicotinic Drugs in the Endoplasmic Reticulum: Beginning the Inside-out Pathway of Addiction and Therapy
J.Gen.Physiol., 2019
|
|
6MDR
 
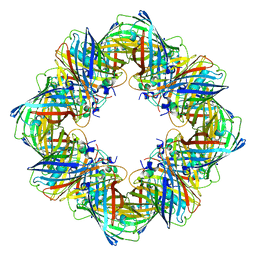 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
6GO9
 
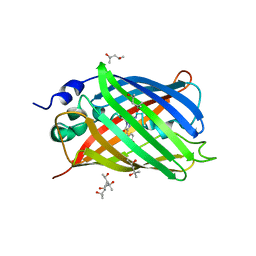 | | Structure of GFPmut2 crystallized at pH 6 and transferred to pH 7 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Lolli, G, Raboni, S, Pasqualetto, E, Campanini, B, Mozzarelli, A, Bettati, S, Battistutta, R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.672 Å) | | Cite: | Insight into GFPmut2 pH Dependence by Single Crystal Microspectrophotometry and X-ray Crystallography.
J.Phys.Chem.B, 122, 2018
|
|
6GO8
 
 | | Structure of GFPmut2 crystallized at pH 6 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Lolli, G, Raboni, S, Pasqualetto, E, Campanini, B, Mozzarelli, A, Bettati, S, Battistutta, R. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Insight into GFPmut2 pH Dependence by Single Crystal Microspectrophotometry and X-ray Crystallography.
J.Phys.Chem.B, 122, 2018
|
|
6GRM
 
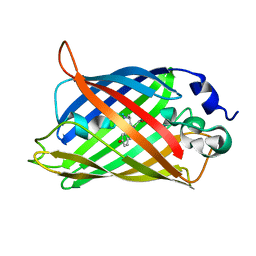 | | Structure of GFPmut2 crystallized at pH 6 and transferred to pH 9 | | Descriptor: | Green fluorescent protein | | Authors: | Lolli, G, Raboni, S, Pasqualetto, E, Campanini, B, Mozzarelli, A, Bettati, S, Battistutta, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into GFPmut2 pH Dependence by Single Crystal Microspectrophotometry and X-ray Crystallography.
J.Phys.Chem.B, 122, 2018
|
|
6GQG
 
 | |
6GQH
 
 | |
6MGH
 
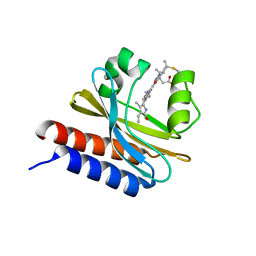 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
6F5J
 
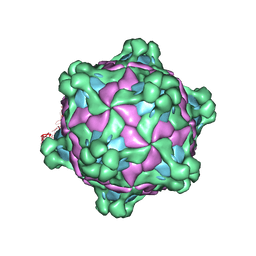 | |
6MWQ
 
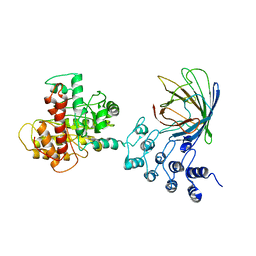 | |
6HWP
 
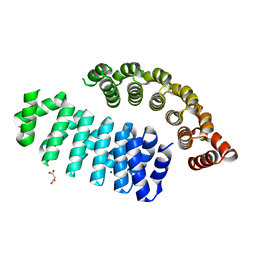 | |
6DQ1
 
 | |
6DQ0
 
 | | sfGFP D133 mutated to 4-nitro-L-phenylalanine | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, superfolder green fluorescent protein | | Authors: | Phillips-Piro, C.M, Maurici, N, Lee, B. | | Deposit date: | 2018-06-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Crystal structures of green fluorescent protein with the unnatural amino acid 4-nitro-L-phenylalanine.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6B9C
 
 | | Superfolder Green Fluorescent Protein with 4-nitro-L-phenylalanine at the chromophore (position 66) | | Descriptor: | CARBON DIOXIDE, Green fluorescent protein | | Authors: | Phillips-Piro, C.M, Brewer, S.H, Olenginski, G.M, Piacentini, J. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
6FP7
 
 | | mTFP1/DARPin 1238_E11 complex in space group P6522 | | Descriptor: | DARPin1238_E11, GFP-like fluorescent chromoprotein cFP484, GLYCEROL | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP8
 
 | | mTFP1/DARPin 1238_E11 complex in space group C2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, DARPin1238_E11, ... | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FP9
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPA
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group P212121 | | Descriptor: | DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6FPB
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
5NHN
 
 | |
5NI3
 
 | | sfGFP 204-204 mutant dimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Worthy, H.L, Rizkallah, P.J. | | Deposit date: | 2017-03-23 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Association of Fluorescent Protein Pairs and Its Significant Impact on Fluorescence and Energy Transfer
Adv Sci, 2020
|
|
6G7P
 
 | | Trichodesmium Tery_3377 (IdiA) (FutA) with iron and water ligands. | | Descriptor: | CHLORIDE ION, Extracellular solute-binding protein, family 1, ... | | Authors: | Machelett, M.M, Tews, I. | | Deposit date: | 2018-04-06 | | Release date: | 2018-09-12 | | Last modified: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of IdiA/FutA (Tery_3377), an iron-binding protein from the ocean diazotrophTrichodesmium erythraeum.
J. Biol. Chem., 293, 2018
|
|
6G7N
 
 | | Trichodesmium Tery_3377 (IdiA) (FutA) with iron and alanine ligand. | | Descriptor: | D-ALANINE, D-GLUTAMIC ACID, Extracellular solute-binding protein, ... | | Authors: | Machelett, M.M, Tews, I. | | Deposit date: | 2018-04-06 | | Release date: | 2018-09-12 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional characterization of IdiA/FutA (Tery_3377), an iron-binding protein from the ocean diazotrophTrichodesmium erythraeum.
J. Biol. Chem., 293, 2018
|
|
6G7Q
 
 | |
6MB2
 
 | | Cryo-EM structure of the PYD filament of AIM2 | | Descriptor: | Green fluorescent protein, Interferon-inducible protein AIM2 | | Authors: | Lu, A, Li, Y, Wu, H. | | Deposit date: | 2018-08-29 | | Release date: | 2018-09-05 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2.
Cell Discov, 1, 2015
|
|
