1R6A
 
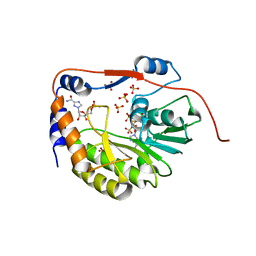 | | Structure of the dengue virus 2'O methyltransferase in complex with s-adenosyl homocysteine and ribavirin 5' triphosphate | | Descriptor: | Genome polyprotein, RIBAVIRIN MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Benarroch, D, Egloff, M.P, Mulard, L, Romette, J.L, Canard, B. | | Deposit date: | 2003-10-15 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate.
J.Biol.Chem., 279, 2004
|
|
1OK4
 
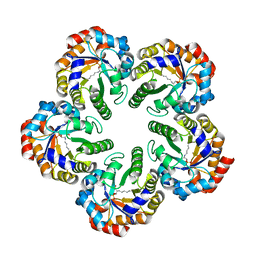 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK6
 
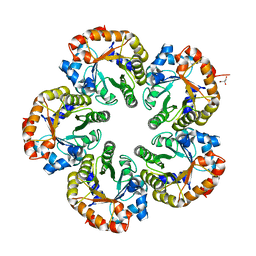 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1RF4
 
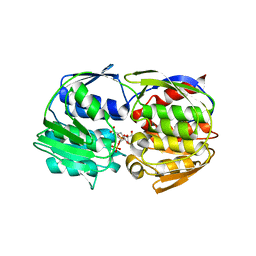 | | Structural Studies of Streptococcus pneumoniae EPSP Synthase, Tetrahedral intermediate Bound State | | Descriptor: | (3R,4S,5R)-5-{[(1R)-1-CARBOXY-2-FLUORO-1-(PHOSPHONOOXY)ETHYL]OXY}-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 5-enolpyruvylshikimate-3-phosphate synthase | | Authors: | Park, H, Hilsenbeck, J.L, Kim, H.J, Shuttleworth, W.A, Park, Y.H, Evans, J.N, Kang, C. | | Deposit date: | 2003-11-07 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state.
Mol.Microbiol., 51, 2004
|
|
2DMY
 
 | | Solution structure of DSRM domain in Spermatid perinuclear RNA-bind protein | | Descriptor: | Spermatid perinuclear RNA-binding protein | | Authors: | Kadirvel, S, He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DSRM domain in Spermatid perinuclear RNA-bind protein
To be Published
|
|
1PC6
 
 | | Structural Genomics, NinB | | Descriptor: | BETA-MERCAPTOETHANOL, Protein ninB | | Authors: | Zhang, R, Beasley, S, Maxwell, K.L, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-15 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1PCM
 
 | |
2DSA
 
 | | Ternary complex of BphK, a bacterial GST | | Descriptor: | (2Z,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, GLUTATHIONE, Glutathione S-transferase | | Authors: | Tocheva, E.I, Murphy, M.E.P. | | Deposit date: | 2006-06-24 | | Release date: | 2006-08-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of ternary complexes of BphK, a bacterial glutathione S-transferase that reductively dechlorinates polychlorinated biphenyl metabolites
J.Biol.Chem., 281, 2006
|
|
1PI3
 
 | | E28Q mutant Benzoylformate Decarboxylase From Pseudomonas Putida | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Bera, A.K, Hasson, M.S. | | Deposit date: | 2003-05-29 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High resolution structure of Benzoylformate Decarboxylate E28Q mutant
To be Published
|
|
1PCJ
 
 | |
2DWD
 
 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DH4
 
 | | Geranylgeranyl pyrophosphate synthase | | Descriptor: | MAGNESIUM ION, YPL069C | | Authors: | Chang, T.-H, Guo, R.-T, Ko, T.-P, Wang, A.H, Liang, P.-H. | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Type-III Geranylgeranyl Pyrophosphate Synthase from Saccharomyces cerevisiae and the Mechanism of Product Chain Length Determination.
J.Biol.Chem., 281, 2006
|
|
1PEW
 
 | | High Resolution Crystal Structure of Jto2, a mutant of the non-amyloidogenic Lamba6 Light Chain, Jto | | Descriptor: | CADMIUM ION, Jto2, a LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN, ... | | Authors: | Dealwis, C, Gupta, V, Wilkerson, M. | | Deposit date: | 2003-05-22 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four V(lambda)6 proteins
J.Mol.Recog., 17, 2004
|
|
2DK6
 
 | | Solution structure of WWE domain in poly (ADP-ribose) polymerase family, member 11 (PARP 11) | | Descriptor: | PARP11 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in poly (ADP-ribose) polymerase family, member 11 (PARP 11)
To be Published
|
|
2DWE
 
 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DK7
 
 | | Solution structure of WW domain in transcription elongation regulator 1 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-06 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WW domain in transcription elongation regulator 1
To be Published
|
|
1PI8
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
1UFN
 
 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
1U1X
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1PIX
 
 | | Crystal structure of the carboxyltransferase subunit of the bacterial ion pump glutaconyl-coenzyme A decarboxylase | | Descriptor: | FORMIC ACID, Glutaconyl-CoA decarboxylase A subunit, SULFATE ION | | Authors: | Wendt, K.S, Schall, I, Huber, R, Buckel, W, Jacob, U. | | Deposit date: | 2003-05-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase
Embo J., 22, 2003
|
|
2BIT
 
 | | Crystal structure of human cyclophilin D at 1.7 A resolution | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2C4X
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
1UFM
 
 | | Solution structure of the PCI domain | | Descriptor: | COP9 complex subunit 4 | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Shirouzu, M, Hayashizaki, Y, The RIKEN Genome Exploration Research Group Phase I & II Teams, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PCI domain
To be Published
|
|
1NP3
 
 | | Crystal structure of class I acetohydroxy acid isomeroreductase from Pseudomonas aeruginosa | | Descriptor: | Ketol-acid reductoisomerase | | Authors: | Ahn, H.J, Eom, S.J, Yoon, H.-J, Lee, B.I, Cho, H, Suh, S.W. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Class I Acetohydroxy Acid Isomeroreductase from Pseudomonas aeruginosa
J.Mol.Biol., 328, 2003
|
|
