6Y1C
 
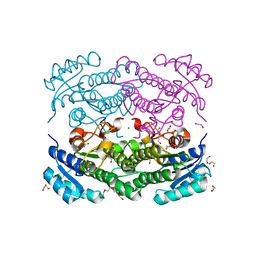 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant D54F | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
9OLN
 
 | |
9SG3
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | Descriptor: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2025-08-21 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Detection and characterisation of ligand-induced conformational changes in acetylcholine binding proteins using biosensors and X-ray crystallography.
Rsc Chem Biol, 6, 2025
|
|
9RCW
 
 | |
9N7B
 
 | |
9R8O
 
 | |
9OTW
 
 | | Influenza A Virus Nucleoprotein(8-498)NP complex with 5-(4-{[(2R)-6,6-dimethyl-1,4-dioxan-2-yl]methoxy}phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide (compound 13) | | Descriptor: | 5-(4-{[(2R)-6,6-dimethyl-1,4-dioxan-2-yl]methoxy}phenyl)-2-oxo-6-(trifluoromethyl)-1,2-dihydropyridine-3-carboxamide, MAGNESIUM ION, Nucleoprotein | | Authors: | Mamo, M. | | Deposit date: | 2025-05-27 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Optimization of a Novel Series of Influenza A Virus Replication Inhibitors Targeting the Nucleoprotein Protein-Protein Interaction.
J.Med.Chem., 68, 2025
|
|
8F88
 
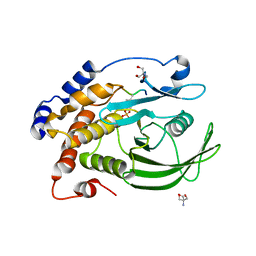 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with monophosphorylated JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
9NNF
 
 | | Cryo-EM structure of a de-novo designed binder NY1-B04 in complex with HLA-A*02:01 and NY-ESO-1-derived peptide SLLMWITQC | | Descriptor: | Beta-2-microglobulin, De-novo designed binder NY1-B04, HLA class I histocompatibility antigen, ... | | Authors: | Gharpure, A, Fernandez-Quintero, M.L, Ward, A.B. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | De novo-designed pMHC binders facilitate T cell-mediated cytotoxicity toward cancer cells.
Science, 389, 2025
|
|
9OJO
 
 | | Crystal structure of TNF alpha in complex with compound 1 | | Descriptor: | 2-{5-[(1S,10S)-1-phenyl-1,2,3,4-tetrahydropyrido[1,2-a][1,3]benzimidazol-8-yl]pyrimidin-2-yl}propan-2-ol, Tumor necrosis factor | | Authors: | Judge, R.A, Bian, Z, Longenecker, K.L. | | Deposit date: | 2025-05-08 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.358 Å) | | Cite: | Discovery of Orally Efficacious Bridged Piperazines as smTNF Modulators.
J.Med.Chem., 68, 2025
|
|
8TXG
 
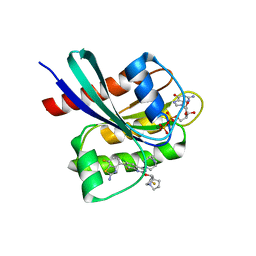 | | Crystal structure of KRAS G12D in complex with GDP and compound 8 | | Descriptor: | (4M)-4-(6-chloro-4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-7-yl)-7-fluoro-1,3-benzothiazol-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, P, Irimia, A, Yang, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Design and Synthesis of Potent and Selective KRAS G12D Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8CLV
 
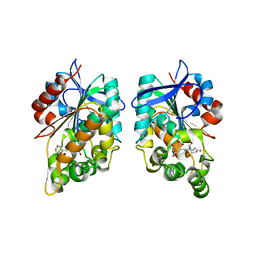 | |
9SKP
 
 | | Crystal structure of HLA-A0201 in complex with peptide SLLWNGPMAV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Liu, Y, Gfeller, D, Guillaume, P, Larabi, A, Lau, K, Pojer, F. | | Deposit date: | 2025-09-02 | | Release date: | 2025-09-17 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of HLA-A0201 in complex with peptide SLLWNGPMAV
To Be Published
|
|
8KGW
 
 | | Molecular mechanism of prostaglandin transporter SLCO2A1 | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Solute carrier organic anion transporter family member 2A1 | | Authors: | Li, Y.H, Qu, Q.H, Zhu, Z.N, Chao, Y.L, Zhou, Z.X. | | Deposit date: | 2023-08-19 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism of prostaglandin transporter SLCO2A1
To Be Published
|
|
8EYB
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2 activation loop phosphopeptide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
7MFH
 
 | | Crystal structure of BIO-32546 bound mouse Autotaxin | | Descriptor: | (1R,3S,5S)-8-{(1S)-1-[8-(trifluoromethyl)-7-{[(1s,4R)-4-(trifluoromethyl)cyclohexyl]oxy}naphthalen-2-yl]ethyl}-8-azabicyclo[3.2.1]octane-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chodaparambil, J.V. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent Selective Nonzinc Binding Autotaxin Inhibitor BIO-32546.
Acs Med.Chem.Lett., 12, 2021
|
|
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6PO4
 
 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
6W1B
 
 | |
5MOS
 
 | | Joint X-ray/neutron structure of cationic trypsin in complex with N-amidinopiperidine | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Schiebel, J, Schrader, T.E, Ostermann, A, Heine, A, Klebe, G. | | Deposit date: | 2016-12-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (0.96 Å), X-RAY DIFFRACTION | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
7OEY
 
 | | Neisseria gonnorhoeae variant E93Q at 1.35 angstrom resolution | | Descriptor: | GLYCEROL, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Widespread occurrence of covalent lysine-cysteine redox switches in proteins.
Nat.Chem.Biol., 18, 2022
|
|
6W2J
 
 | | CPS1 bound to allosteric inhibitor H3B-374 | | Descriptor: | (2-fluoranyl-4-methoxy-phenyl)-[(3~{R},5~{R})-4-(2-fluoranyl-4-methoxy-phenyl)carbonyl-3,5-dimethyl-piperazin-1-yl]methanone, 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2020-03-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of 2,6-Dimethylpiperazines as Allosteric Inhibitors of CPS1.
Acs Med.Chem.Lett., 11, 2020
|
|
5FXV
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with N05859b | | Descriptor: | 1,2-ETHANEDIOL, 4-CARBOXYPIPERIDINE, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Srikannathasan, V, Oerum, S, Pearce, N.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with N05859B
To be Published
|
|
5FXW
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Fumarate
To be Published
|
|
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
