8VHQ
 
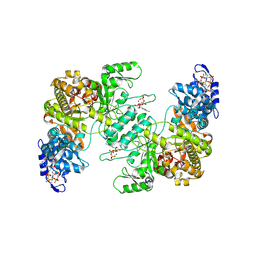 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to dATP and ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHP
 
 | | Crystal structure of E. coli class Ia ribonucleotide reductase alpha subunit W28A variant bound to CDP and two molecules of ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHO
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHN
 
 | | Crystal Structure of E. coli class Ia ribonucleotide reductase alpha subunit bound to two ATP molecules | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8VHM
 
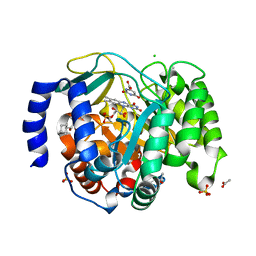 | | Structure of DHODH in Complex with Fragment 2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VHL
 
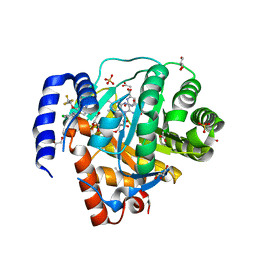 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8VHH
 
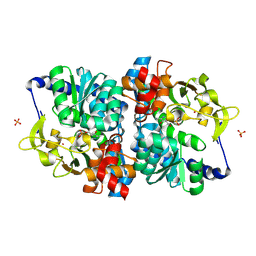 | | Engineered holo tryptophan synthase (Tm9D8*) derived from T. maritima TrpB | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Porter, N.J, Johnston, K.E, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2024-01-02 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A combinatorially complete epistatic fitness landscape in an enzyme active site.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VHF
 
 | | Cryo-EM structure of GPR119-Gs-Nb35 complex with small molecule agonist MBX-2982 | | Descriptor: | 2-[1-(5-ethylpyrimidin-2-yl)piperidin-4-yl]-4-[[4-(1,2,3,4-tetrazol-1-yl)phenoxy]methyl]-1,3-thiazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kim, D.-G, Cherezov, V. | | Deposit date: | 2024-01-01 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Native mass spectrometry prescreening of G protein-coupled receptor complexes for cryo-EM structure determination
To Be Published
|
|
8VHE
 
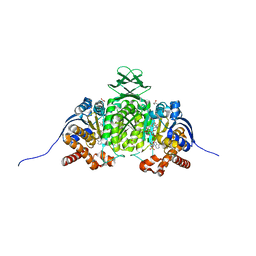 | | Crystal Structure of Human IDH1 R132Q in Complex with NADPH-TCEP Adduct | | Descriptor: | 3,3',3''-({(4R)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1,4-dihydropyridin-4-yl}-lambda~5~-phosphanetriyl)tripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHD
 
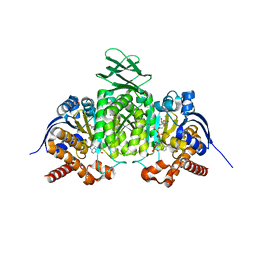 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Isocitrate | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHC
 
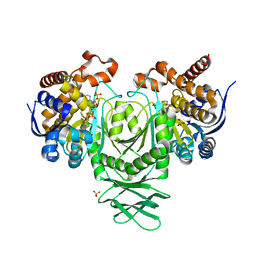 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHB
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | Descriptor: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHA
 
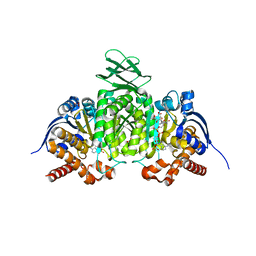 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | Descriptor: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VH9
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH | | Descriptor: | CITRIC ACID, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Mealka, M, Sohl, C.D, Huxford, T. | | Deposit date: | 2023-12-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VH8
 
 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 2.85 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
8VH7
 
 | | Crystal structure of heparosan synthase 2 from Pasteurella multocida at 1.98 A | | Descriptor: | 1,2-ETHANEDIOL, Heparosan synthase B, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Liu, J, Stancanelli, E, Krahn, J.M. | | Deposit date: | 2023-12-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
8VH5
 
 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH4
 
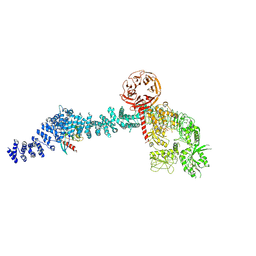 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH3
 
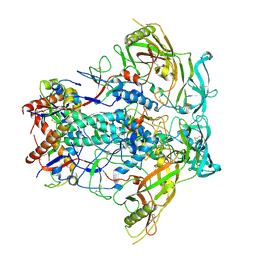 | | CH505.M5.G458Y CE2 Design SOSIP | | Descriptor: | CH505.CE2 SOSIP gp120, CH505.CE2 SOSIP gp41 | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VH2
 
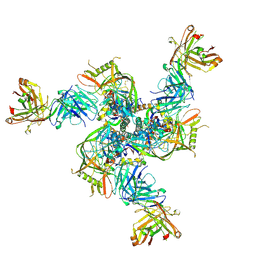 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.12 Fab Heavy Chain, CH235.12 Fab Light Chain, CH505.M5.G458Y SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VH1
 
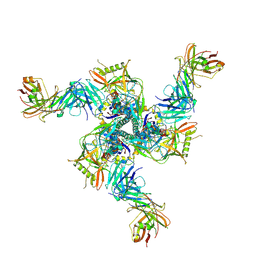 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.I60 Fab Heavy Chain, CH235.I60 Fab Light Chain, CH505.M5 SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VGZ
 
 | |
8VGW
 
 | | VRC01 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, VRC01 Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VGV
 
 | | DH270.6 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, DH270.6 Antibody Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VGT
 
 | |
