7QS1
 
 | |
7QRV
 
 | | Crystal structure of NHL domain of TRIM2 (full C-terminal) | | Descriptor: | 1,2-ETHANEDIOL, Tripartite motif-containing protein 2 | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Comparative structural analyses of the NHL domains from the human E3 ligase TRIM-NHL family.
Iucrj, 9, 2022
|
|
8Y56
 
 | | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS | | Descriptor: | CHOLESTEROL, Lysosomal cholesterol signaling protein, SODIUM ION, ... | | Authors: | Zhao, J, Shen, Q.Y, Zhang, Y, Shao, Z.H. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM reveals cholesterol binding in the lysosomal GPCR-like protein LYCHOS.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6VN3
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 23 | | Descriptor: | 1-{[7-(5-chloro-2-{[(3R,4S)-4-fluoropyrrolidin-3-yl]oxy}-3-methylphenyl)thieno[3,2-b]pyridin-2-yl]methyl}-1H-pyrrole-2,5-dione, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
7QRX
 
 | |
7QT6
 
 | | Room temperature In-situ SARS-CoV-2 MPRO with bound Z1367324110 | | Descriptor: | 1-methyl-3,4-dihydro-2~{H}-quinoline-7-sulfonamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Horrell, S, Gildae, R.J, Axford, D, Owen, C.D, Lukacik, P, Strain-Damerell, C, Owen, R.L, Walsh, M.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | xia2.multiplex: a multi-crystal data-analysis pipeline.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6FUP
 
 | |
8EER
 
 | |
7OKA
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 14 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-(1~{H}-imidazol-4-ylmethyl)ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
8EI8
 
 | | Crystal structure of the WWP2 HECT domain in complex with H308, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, H308, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
7OJQ
 
 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 7 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-methyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7OEF
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ038 | | Descriptor: | 4-(2-azaspiro[3.4]octan-2-ylmethyl)-~{N}-[[(3~{R})-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
9FBZ
 
 | | Dye-decolourising peroxidase DtpB mixed with hydrogen peroxide for 1.3 s | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Owen, R.L, Devenish, N. | | Deposit date: | 2024-05-15 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Dye-decolourising peroxidase DtpB mixed with hydrogen peroxide for 1.3 s
To Be Published
|
|
8CIQ
 
 | | JzTx-34 toxin peptide | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJP
 
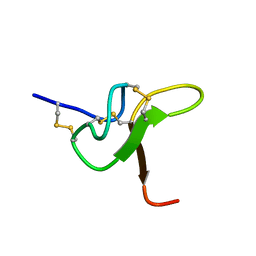 | | JzTx-34 toxin peptide H18A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
7OJW
 
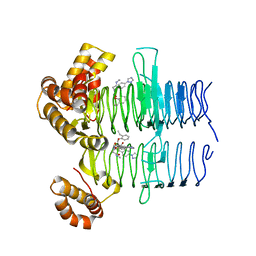 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 93 | | Descriptor: | 2-[2-(2-ethylphenoxy)ethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-~{N}-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
8SPV
 
 | | PS3 F1 Rotorless, no ATP | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
8CJS
 
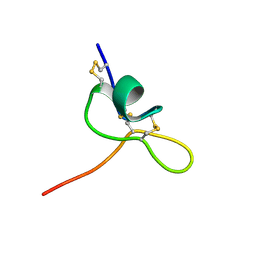 | | JzTx-34 toxin peptide W31A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJQ
 
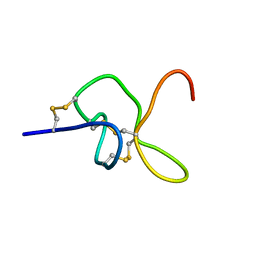 | | JzTx-34 toxin peptide E20A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
6PO4
 
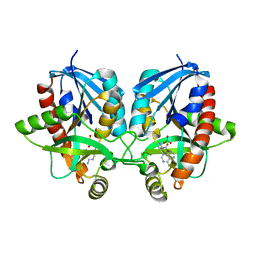 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
8CJR
 
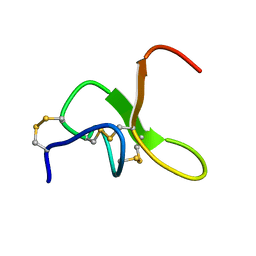 | | JzTx-34 toxin peptide W25A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8CJT
 
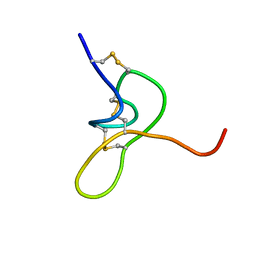 | | JzTx-34 toxin peptide W33A mutant | | Descriptor: | Mu-theraphotoxin-Cg1a | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2023-02-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of new peptides activating human Na v 1.1.
Biomed Pharmacother, 165, 2023
|
|
8EJV
 
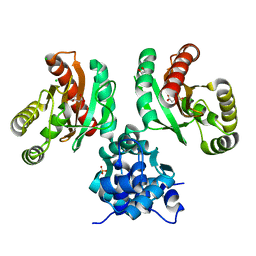 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and structural characterization of an IclR family transcription factor for the development of dicarboxylic acid biosensors.
Febs J., 291, 2024
|
|
7NX1
 
 | | TG domain of LTK | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leukocyte tyrosine kinase receptor, TERBIUM(III) ION | | Authors: | De Munck, S, Savvides, S.N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cytokine-mediated activation of ALK family receptors.
Nature, 600, 2021
|
|
