1J6W
 
 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J7I
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa Apoenzyme | | Descriptor: | AMINOGLYCOSIDE 3'-PHOSPHOTRANSFERASE | | Authors: | Burk, D.L, Hon, W.C, Leung, A.K.-W, Berghuis, A.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analyses of nucleotide binding to an aminoglycoside phosphotransferase.
Biochemistry, 40, 2001
|
|
4UFN
 
 | | Laboratory evolved variant R-C1B1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EPOXIDE HYDROLASE | | Authors: | Carlsson, A.J, Bauer, P, Nilsson, M, Dobritzsch, D, Kamerlin, S.C.L, Widersten, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Diversity and Enantioconvergence in Potato Epoxide Hydrolase 1.
Org.Biomol.Chem., 14, 2016
|
|
8RK0
 
 | | HCV E1/E2 homodimer complex, ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HCV E1, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
8RJJ
 
 | | HCV E1/E2 homodimer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
5LD9
 
 | | Structure of deubiquitinating enzyme homolog, Pyrococcus furiosus JAMM1. | | Descriptor: | CHLORIDE ION, JAMM1, ZINC ION | | Authors: | Maupin-Furlow, J.A, Franzetti, B, Cao, S, Girard, E, Gabel, F, Engilberge, S. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-17 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
8R8E
 
 | | DYRK1a in Complex with 2-Cyclopentyl-7-iodo-1H-indole-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-7-iodanyl-1~{H}-indole-3-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Dammann, M, Stehle, T, Boeckler, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Halogen Bonding on Water─A Drop in the Ocean?
J Chem Theory Comput, 2024
|
|
1ZU2
 
 | |
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
4W2H
 
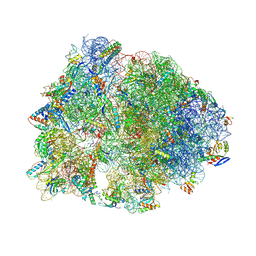 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
2IZ1
 
 | | 6PDH complexed with PEX inhibitor synchrotron data | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-23 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Bacterial 6- Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
2IZ0
 
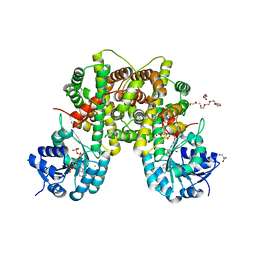 | | PEX inhibitor-home data | | Descriptor: | 1,2-ETHANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Sundaramoorthy, R, Iulek, J, Hunter, W.N. | | Deposit date: | 2006-07-23 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of a Bacterial 6- Phosphogluconate Dehydrogenase Reveal Aspects of Specificity, Mechanism and Mode of Inhibition by Analogues of High-Energy Reaction Intermediates.
FEBS J., 274, 2007
|
|
8TI6
 
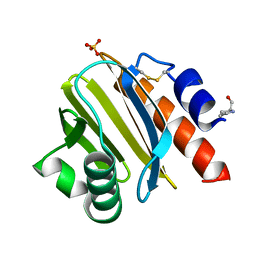 | | Crystal structure of Tyr p 36.0101 | | Descriptor: | Profilin, Proline-rich peptide, SULFATE ION | | Authors: | O'Malley, A, Sankaran, S, Chruszcz, M. | | Deposit date: | 2023-07-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural homology of mite profilins to plant profilins is not indicative of allergic cross-reactivity.
Biol.Chem., 405, 2024
|
|
8TI5
 
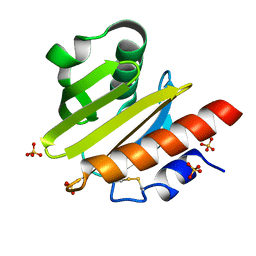 | | Crystal structure of Tyr p 36.0101 | | Descriptor: | Profilin, SULFATE ION | | Authors: | O'Malley, A, Sankaran, S, Chruszcz, M. | | Deposit date: | 2023-07-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural homology of mite profilins to plant profilins is not indicative of allergic cross-reactivity.
Biol.Chem., 405, 2024
|
|
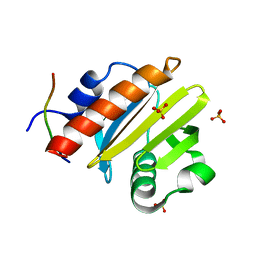
8TI7
 
 | |
3K7W
 
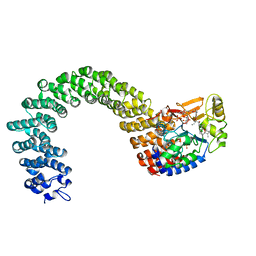 | | Protein phosphatase 2A core complex bound to dinophysistoxin-2 | | Descriptor: | (2R)-2-hydroxy-3-[(2S,5R,6R,8S)-5-hydroxy-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-8'-hydroxy-6'-{(1S,3S)-1-hydroxy-3-[( 2S,6R,11S)-11-methyl-1,7-dioxaspiro[5.5]undec-2-yl]butyl}-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]p yran]-5-yl]-1-methylprop-2-en-1-yl}-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
3K7V
 
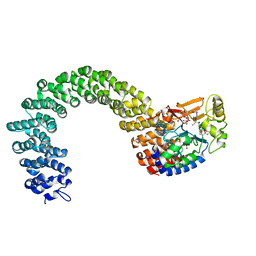 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | Descriptor: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
6ZPP
 
 | |
5KQJ
 
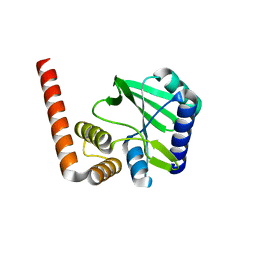 | |
5LDA
 
 | | Structure of deubiquitinating enzyme homolog (Pyrococcus furiosus JAMM1) in complex with ubiquitin-like SAMP2. | | Descriptor: | GLYCEROL, JAMM1, SAMP2, ... | | Authors: | Cao, S, Engilberge, S, Girard, E, Gabel, F, Franzetti, B, Maupin-Furlow, J.A. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
5MJL
 
 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
8V5Y
 
 | |
2B08
 
 | | Reduced acetamide-bound M150G Nitrite Reductase from Alcaligenes faecalis | | Descriptor: | ACETAMIDE, COPPER (I) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, MacPherson, I.S, Farver, O, Tocheva, E.I, Pecht, I, Verbeet, M.Ph, Murphy, M.E.P, Canters, G.W. | | Deposit date: | 2005-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of the methionine ligand on the reorganization energy of the type-1 copper site of nitrite reductase.
J.Am.Chem.Soc., 129, 2007
|
|
2JON
 
 | | Solution structure of the C-terminal domain Ole e 9 | | Descriptor: | Beta-1,3-glucanase | | Authors: | Trevino, M.A, Palomares, O, Castrillo, I, Villalba, M, Rodriguez, R, Rico, M, Santoro, J, Bruix, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ole e 9, a major allergen of olive pollen
Protein Sci., 17, 2008
|
|
4KEF
 
 | |
