7ON5
 
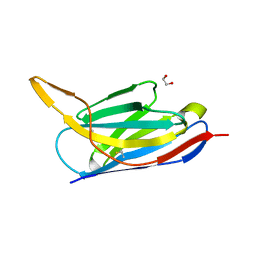 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
4USC
 
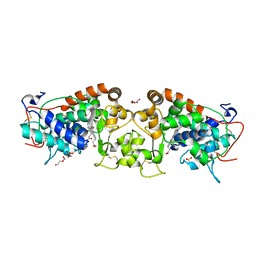 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
3N4O
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*(B7C)P*CP*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S. | | Deposit date: | 2010-05-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
4UFB
 
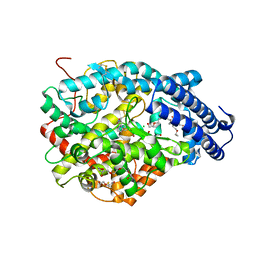 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with Lys-Pro | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Masuyer, G, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ac-Sdkp Hydrolysis by Angiotensin-I Converting Enzyme
Sci.Rep., 5, 2015
|
|
4UFA
 
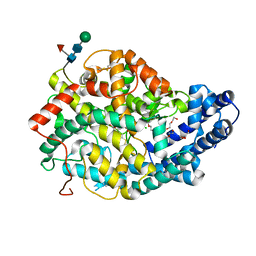 | | Crystal structure of the Angiotensin-1 converting enzyme N-domain in complex with Ac-SD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, ... | | Authors: | Masuyer, G, Douglas, R.G, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ac-Sdkp Hydrolysis by Angiotensin-I Converting Enzyme
Sci.Rep., 5, 2015
|
|
4V3U
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(pyridin-3-yl) propan-1-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}-3-(pyridin-3-yl)propan-1-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
8WNW
 
 | | the structure of PsaQ | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, PsaQ | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
9XZT
 
 | | Crystal structure of BBn6 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, BBn6, SULFATE ION, ... | | Authors: | Bera, A.K, Kim, D, Baker, D. | | Deposit date: | 2025-08-27 | | Release date: | 2025-09-03 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Parametrically guided design of beta barrels and transmembrane nanopores using deep learning.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9UDY
 
 | |
4UO3
 
 | | Structure of the A_Equine_Richmond_07 H3 haemagglutinin mutant Ser30Thr | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vachieri, S.G, Collins, P.J, Haire, L.F, Ogrodowicz, R.W, Martin, S.R, Walker, P.A, Xiong, X, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Recent Evolution of Equine Influenza and the Origin of Canine Influenza.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8F1S
 
 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in A-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8F1V
 
 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in B-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
8FDQ
 
 | |
447D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Squire, C.J, Baker, L.J, Clark, G.R, Martin, R.F, White, J. | | Deposit date: | 1999-01-18 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
4W4O
 
 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
1G4Q
 
 | | RNA/DNA HYBRID DECAMER OF CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', ... | | Authors: | Han, G.W. | | Deposit date: | 2000-10-27 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct-methods determination of an RNA/DNA hybrid decamer at 1.15 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
9PWN
 
 | | Crystal structure of Fabs 7411 in complex with TREM2 peptide | | Descriptor: | 1,2-ETHANEDIOL, 7411 Fab Heavy Chain, 7411 Fab Light Chain, ... | | Authors: | Arndt, J.W, Chao, Q, Cooke, H.A, Almeida, A.D.S. | | Deposit date: | 2025-08-04 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Building a potent TREM2 agonistic, biparatopic, common light chain antibody.
Mabs, 17, 2025
|
|
9U3N
 
 | |
9XY2
 
 | | One of a series of engineered variants of I-OnuI meganuclease targeting altered DNA target sequence | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Werther, R, Schwensen, J.S, Stoddard, B.L. | | Deposit date: | 2025-08-25 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Prediction, modeling and design of protein-DNA recognition via computational and machine learning approaches:
accomplishments and remaining challenges
To Be Published
|
|
9NZ3
 
 | | SARS-CoV M protein dimer in complex with JNJ-9676 and FAb B | | Descriptor: | (6S,8R)-N-(3-cyanophenyl)-5-{4-[difluoro(phenyl)methyl]phenyl}-6-methyl-4-oxo-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-3-carboxamide, FAb B heavy chain, FAb B light chain, ... | | Authors: | Mann, M.K, Abeywickrema, P. | | Deposit date: | 2025-03-31 | | Release date: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | SARS-CoV M protein dimer in complex with JNJ-9676 and FAb B
To Be Published
|
|
9XYA
 
 | |
6W47
 
 | | Peptoid-Containing Collagen Peptide | | Descriptor: | 1,2-ETHANEDIOL, Collagen-like peptide | | Authors: | Chenoweth, D.M, Melton, S.D. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Rules for the design of aza-glycine stabilized triple-helical collagen peptides.
Chem Sci, 11, 2020
|
|
6VZC
 
 | |
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9LE3
 
 | | Consensus map of UCC118 Rool RNA | | Descriptor: | UCC118 Rool RNA | | Authors: | Zhang, K, Li, S. | | Deposit date: | 2025-01-07 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural insights into higher-order natural RNA-only multimers.
Nat.Struct.Mol.Biol., 32, 2025
|
|
