6SYA
 
 | |
6MRM
 
 | | Red Clover Necrotic Mosaic Virus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
5C7P
 
 | |
6G4N
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(oxidanylamino)methyl]pyridin-3-ol, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6MRL
 
 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
6MS2
 
 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | Descriptor: | CALCIUM ION, Glycoside Hydrolase Family 43 | | Authors: | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | Deposit date: | 2018-10-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
6G17
 
 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by nerve agent tabun | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6G4P
 
 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(~{Z})-hydroxyiminomethyl]pyridin-3-ol, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
5CAA
 
 | |
4RAK
 
 | |
4RA8
 
 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
6NL3
 
 | | Solution structure of human Coa6 | | Descriptor: | Cytochrome c oxidase assembly factor 6 homolog | | Authors: | Naik, M.T, Soma, S, Gohil, V. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase.
Cell Rep, 29, 2019
|
|
7C5D
 
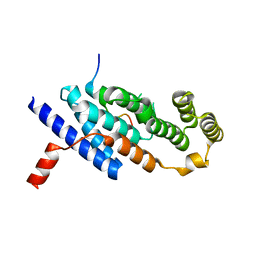 | | Crystal structure of TRF2 TRFH domain in complex with a MCPH1 peptide | | Descriptor: | GLYCEROL, Microcephalin, Telomeric repeat-binding factor 2 | | Authors: | Xiong, X, Chen, Y. | | Deposit date: | 2020-05-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Microcephalin 1/BRIT1-TRF2 interaction promotes telomere replication and repair, linking telomere dysfunction to primary microcephaly.
Nat Commun, 11, 2020
|
|
4RHZ
 
 | | Crystal structure of Cry23Aa1 and Cry37Aa1 binary protein complex | | Descriptor: | CALCIUM ION, Cry23AA1, Cry37AA1, ... | | Authors: | Rydel, T.J, Williams, J.M, Brown, G.R, Guzov, V.M, Sturman, E.J, Evdokimov, A. | | Deposit date: | 2014-10-03 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Associated Bacillus thuringiensis Binary Protein Complex of Cry23Aa1 and Cry37Aa1: Crystal Structure, Insecticidal Data, and Pore Formation Modeling.
To be Published
|
|
6H6A
 
 | | Crystal structure of UNC119 in complex with LCK peptide | | Descriptor: | GLY-CYS-GLY-CYS-SER-SER, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Yelland, T, ElMaghloob, Y, McIlwraith, M, Stephen, L, Ismail, S. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-26 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK.
Dev.Cell, 47, 2018
|
|
5O4L
 
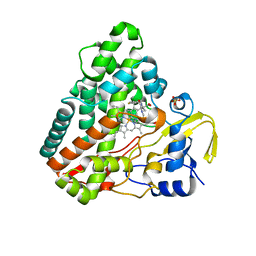 | | Crystal structure of P450 CYP121 in complex with compound 6a. | | Descriptor: | 1-[(4-fluorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
6U6Z
 
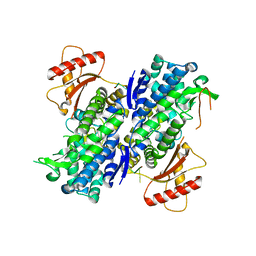 | | Human SAMHD1 bound to deoxyribo(TG*TTCA)-oligonucleotide | | Descriptor: | DNA polymer TG(PST)TCA, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Yu, C.H, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
1QIV
 
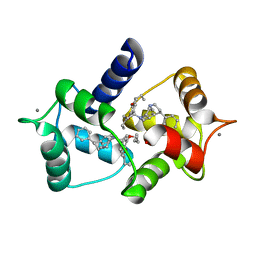 | | CALMODULIN COMPLEXED WITH N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE (DPD), 1:2 COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Naray-Szabo, G, Ovadi, J. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
1QIW
 
 | | Calmodulin complexed with N-(3,3,-diphenylpropyl)-N'-[1-R-(3,4-bis-butoxyphenyl)-ethyl]-propylenediamine (DPD) | | Descriptor: | CALCIUM ION, CALMODULIN, N-(3,3,-DIPHENYLPROPYL)-N'-[1-R-(2 3,4-BIS-BUTOXYPHENYL)-ETHYL]-PROPYLENEDIAMINE | | Authors: | Harmat, V, Bocskei, Z.S, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1999-06-17 | | Release date: | 2000-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Potent Calmodulin Antagonist with Arylalkylamine Structure: Crystallographic, Spectroscopic and Functional Studies
J.Mol.Biol., 297, 2000
|
|
8IZD
 
 | | Cryo-EM structure of the C26-CoA-bound Lac1-Lip1 complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1, ... | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
6H5H
 
 | |
6NZC
 
 | |
