6ZT7
 
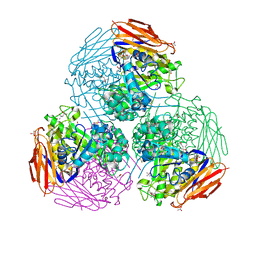 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
5F4O
 
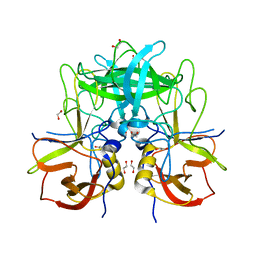 | |
7A8O
 
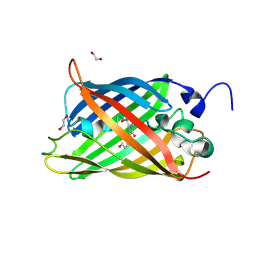 | |
7AA0
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | (~{E})-3-[4-(4,4-dimethyl-1-propan-2-yl-2,3-dihydroquinolin-6-yl)phenyl]prop-2-enoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1E4L
 
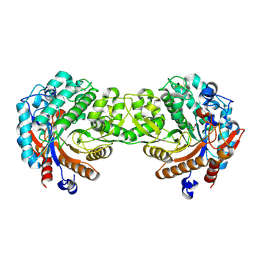 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZM Glu191Asp | | Descriptor: | BETA-GLUCOSIDASE, CHLOROPLASTIC, GLYCEROL | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-10 | | Release date: | 2000-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4FSN
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 4-(6-{[(4-METHYLCYCLOHEXYL)AMINO]METHYL}-1,4-DIHYDROINDENO[1,2-C]PYRAZOL-3-YL)BENZOIC ACID, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
7AAI
 
 | | Crystal structure of Human serum albumin in complex with perfluorooctanoic acid (PFOA) at 2.10 Angstrom Resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Maso, L, Liberi, S, Trande, M, Angelini, A, Cendron, L. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unveiling the binding mode of perfluorooctanoic acid to human serum albumin.
Protein Sci., 30, 2021
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
6NHQ
 
 | |
5LRE
 
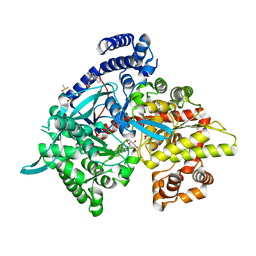 | | Crystal structure of Glycogen Phosphorylase b in complex with KS382 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-31 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic, enzyme kinetic, and protein crystallographic studies of C-beta-d-glucopyranosyl pyrroles and imidazoles reveal and explain low nanomolar inhibition of human liver glycogen phosphorylase.
Eur J Med Chem, 123, 2016
|
|
6ZT4
 
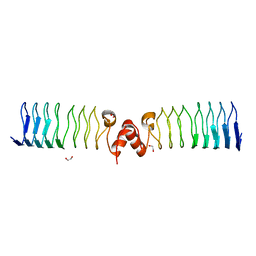 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1E3X
 
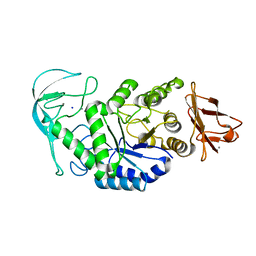 | | Native structure of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.92A | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | Deposit date: | 2000-06-26 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
7A6X
 
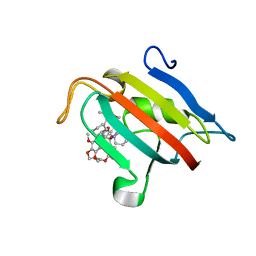 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | Descriptor: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6ZWJ
 
 | | Neisseria gonorrhoeae transaldolase at 1.35 Angstrom resolution | | Descriptor: | CITRIC ACID, GLYCEROL, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
4GCY
 
 | | Structure of Mycobacterium tuberculosis dUTPase H21W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, J, Vertessy, B.G, Leveles, I, Bendes, A. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | RAMD identification of substrate binding pathways to the active site of dUTPase
To be Published
|
|
6ZZ5
 
 | | Structure of soluble SmhB of the tripartite alpha-pore forming toxin, Smh, from Serratia marcescens. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SmhB | | Authors: | Churchill-Angus, A.M, Baker, P.J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterisation of a tripartite alpha-pore forming toxin from Serratia marcescens
Sci Rep, 11, 2021
|
|
6SZG
 
 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GR839 and GSK513 | | Descriptor: | (4-chlorophenyl)-[(3~{S})-3-oxidanylpiperidin-1-yl]methanone, 4,5,6,7-tetrahydro-2~{H}-indazole-3-carboxylic acid, CALCIUM ION, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4GBD
 
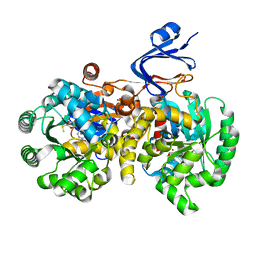 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
7A4T
 
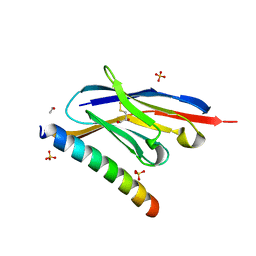 | | Crystal structure of the GCN coiled-coil in complex with nanobody Nb39 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, GCN4 isoform 1, ... | | Authors: | Hadzi, S. | | Deposit date: | 2020-08-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | A nanobody toolbox targeting dimeric coiled-coil modules for functionalization of designed protein origami structures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5LMR
 
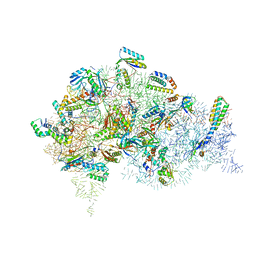 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
6ZOD
 
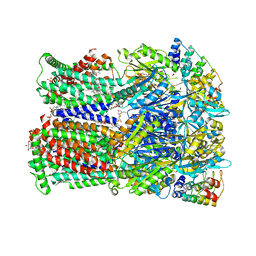 | | Fusidic acid binding to the allosteric deep transmembrane domain binding pocket, TM7/TM8 groove, and TM1/TM2 groove of the fully induced AcrB T protomer | | Descriptor: | 1,2-ETHANEDIOL, DARPIN, DECANE, ... | | Authors: | Oswald, C, Tam, H.K, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
4FV5
 
 | | Crystal Structure of the ERK2 complexed with EK9 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, N-[(1S)-2-hydroxy-1-phenylethyl]-4-(5-methyl-2-phenylpyrimidin-4-yl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the ERK2 complexed with EK9
TO BE PUBLISHED
|
|
4GM0
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403N | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
6NHR
 
 | |
4FV8
 
 | | Crystal Structure of the ERK2 complexed with E63 | | Descriptor: | 1,2-ETHANEDIOL, 6-({4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]-5-(phenylamino)pyrimidin-2-yl}amino)-1,2-dihydro-3H-indazol-3-one, Mitogen-activated protein kinase 1, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Xie, X. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the ERK2 complexed with E63
TO BE PUBLISHED
|
|
