5MXE
 
 | | Photorhabdus asymbiotica lectin (PHL) in free form | | Descriptor: | CHLORIDE ION, Photorhabdus asymbiotica lectin PHL, SODIUM ION | | Authors: | Jancarikova, G, Houser, J, Demo, G, Wimmerova, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of novel bangle lectin from Photorhabdus asymbiotica with dual sugar-binding specificity and its effect on host immunity.
PLoS Pathog., 13, 2017
|
|
5WOR
 
 | |
5MXH
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with D-galactose | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Photorhabdus asymbiotica lectin PHL, ... | | Authors: | Jancarikova, G, Houser, J, Demo, G, Wimmerova, M. | | Deposit date: | 2017-01-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of novel bangle lectin from Photorhabdus asymbiotica with dual sugar-binding specificity and its effect on host immunity.
PLoS Pathog., 13, 2017
|
|
6OWV
 
 | | Crystal structure of a Human Cardiac Calsequestrin Filament | | Descriptor: | CHLORIDE ION, Calsequestrin-2, SULFATE ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OTF
 
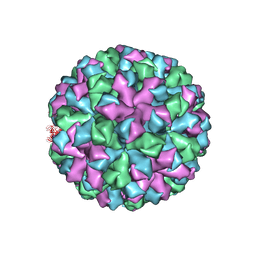 | | Symmetric reconstruction of human norovirus GII.2 Snow Mountain Virus Strain VLP in T=3 symmetry | | Descriptor: | Viral protein 1, ZINC ION | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-03 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OWW
 
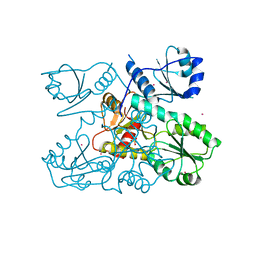 | | Crystal structure of a Human Cardiac Calsequestrin Filament Complexed with Ytterbium | | Descriptor: | Calsequestrin-2, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XKX
 
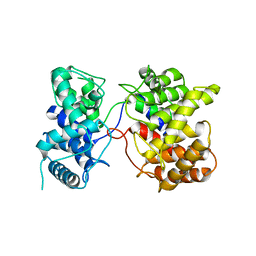 | | Crystal structure of Tpn2 | | Descriptor: | SQHop_cyclase_C domain-containing protein | | Authors: | Chang, C.Y, Stowell, E.A, Lin, Y.L, Ehrenberger, M.A, Rudolf, J.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure-guided product determination of the bacterial type II diterpene synthase Tpn2.
Commun Chem, 5, 2022
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
4YVM
 
 | |
7X6H
 
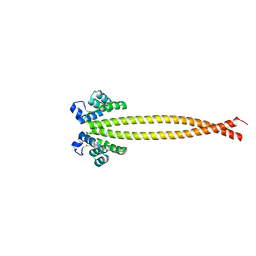 | |
7X6F
 
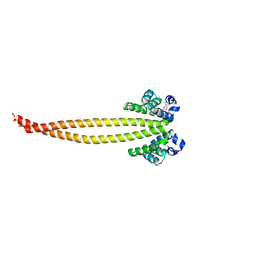 | | Outer membrane lipoprotein QseG of Escherichia coli O157:H7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CITRATE ANION, ... | | Authors: | Matsumoto, K, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-03-07 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
4Y6X
 
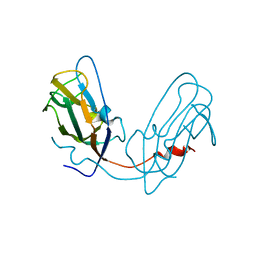 | |
6LF5
 
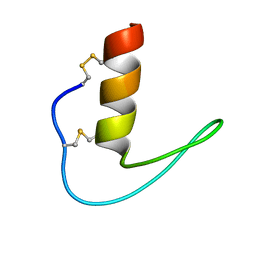 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
7X6G
 
 | |
4YHJ
 
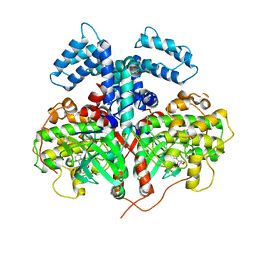 | | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4 (GRK4) | | Descriptor: | AMP PHOSPHORAMIDATE, G protein-coupled receptor kinase 4 | | Authors: | Allen, S.J, Parthasarathy, G, Soisson, S, Munshi, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4.
J.Biol.Chem., 290, 2015
|
|
6ZKY
 
 | | Crystal structure of InhA:01 TCR in complex with HLA-E (S147C) bound to InhA (53-61 H3C) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6C2Z
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6M9C
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR Pseudotyrostatin | | Descriptor: | ACETIC ACID, CALCIUM ION, Pseudotyrostatin, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-23 | | Release date: | 2018-10-24 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
6ZXJ
 
 | | Fully-loaded anthrax lethal toxin in its heptameric pre-pore state, in which the third lethal factor is masked out (PA7LF3-masked) | | Descriptor: | Lethal factor, Protective antigen | | Authors: | Quentin, D, Antoni, C, Gatsogiannis, C, Raunser, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Plos Pathog., 16, 2020
|
|
6M9R
 
 | |
6BPQ
 
 | | Structure of the cold- and menthol-sensing ion channel TRPM8 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Wu, M, Zubcevic, L, Borschel, W.F, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2017-11-25 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the cold- and menthol-sensing ion channel TRPM8.
Science, 359, 2018
|
|
6PIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6C4A
 
 | | Crystal structure of 3-nitropropionate modified isocitrate lyase from Mycobacterium tuberculosis with pyruvate | | Descriptor: | 3-NITROPROPANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kreitler, D.F, Ray, S, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nitro Group as a Masked Electrophile in Covalent Enzyme Inhibition.
ACS Chem. Biol., 13, 2018
|
|
5WLI
 
 | | Crystal Structure of H-2Db with the GAP501 peptide (SQL) | | Descriptor: | Beta-2-microglobulin, GAP50 peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Farenc, C, Josephs, T, Rossjohn, J. | | Deposit date: | 2017-07-27 | | Release date: | 2017-11-15 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A T Cell Receptor Locus Harbors a Malaria-Specific Immune Response Gene.
Immunity, 47, 2017
|
|
