1KVW
 
 | |
4V0S
 
 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
8VZG
 
 | | Carbonic anhydrase II mutant with DRD function | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CITRATE ANION, Carbonic anhydrase 2, ... | | Authors: | Sethi, D.K. | | Deposit date: | 2024-02-11 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Functional regulation of membrane-bound cytokine activity in engineered T cells via drug-responsive domains derived from carbonic anhydrase 2
To Be Published
|
|
8VG9
 
 | |
8VFQ
 
 | | De novo design apixaban-binding protein: apx1049 | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, De novo designed apixaban-binding protein apx1049 | | Authors: | Bera, A.K, Lee, G.R, Baker, D. | | Deposit date: | 2023-12-21 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-molecule binding and sensing with a designed protein family
To Be Published
|
|
1KTU
 
 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1KJ5
 
 | | Solution Structure of Human beta-defensin 1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
5IDY
 
 | |
1KVM
 
 | | X-ray Crystal Structure of AmpC WT beta-Lactamase in Complex with Covalently Bound Cephalothin | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
1KVY
 
 | |
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
8VYS
 
 | | Cryo-EM Structure of the BRAF V600E monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYU
 
 | | Cryo-EM Structure of the BRAF WT monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
5VX0
 
 | | Bak in complex with Bim-h3Glg | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 11, ... | | Authors: | Brouwer, J.M, Lan, P, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VRG
 
 | |
2BMO
 
 | | The Crystal Structure of Nitrobenzene Dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, FE (III) ION, ... | | Authors: | Friemann, R, Ivkovic-Jensen, M.M, Lessner, D.J, Yu, C, Gibson, D.T, Parales, R.E, Eklund, H, Ramaswamy, S. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase.
J. Mol. Biol., 348, 2005
|
|
5VQK
 
 | |
5ID7
 
 | | Crystal structure of human serum albumin in complex with phosphorodithioate derivative of myristoyl cyclic phosphatidic acid (cPA) | | Descriptor: | (4S)-2-sulfanylidene-4-[(tetradecanoyloxy)methyl]-1,3,2lambda~5~-dioxaphospholane-2-thiolate, DI(HYDROXYETHYL)ETHER, Serum albumin, ... | | Authors: | Sekula, B, Bujacz, A, Rytczak, P, Bujacz, G. | | Deposit date: | 2016-02-24 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural evidence of the species-dependent albumin binding of the modified cyclic phosphatidic acid with cytotoxic properties.
Biosci.Rep., 36, 2016
|
|
5IDP
 
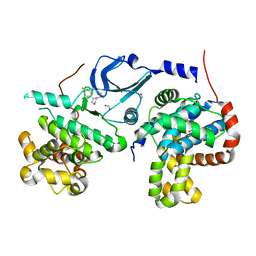 | | CDK8-CYCC IN COMPLEX WITH (3-Amino-1H-indazol-5-yl)-[(S)-2-(4-fluoro-phenyl)-piperidin-1-yl]-methanone | | Descriptor: | (3-amino-1H-indazol-5-yl)[(2S)-2-(4-fluorophenyl)piperidin-1-yl]methanone, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
8VIG
 
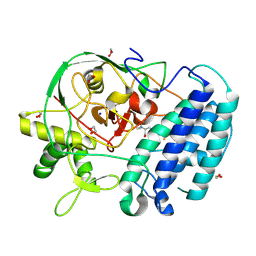 | | EgtB-IV from Geminocystis sp. isolate SKYG4, an ergothioneine-biosynthetic type IV sulfoxide synthase in complex with hercynine | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N,N,N-trimethyl-histidine, ... | | Authors: | Ireland, K.A, Davis, K.M. | | Deposit date: | 2024-01-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the convergent evolution of sulfoxide synthase EgtB-IV, an ergothioneine-biosynthetic homolog of ovothiol synthase OvoA.
Structure, 32, 2024
|
|
5IDU
 
 | |
8VDD
 
 | | Crystal structure of Proinsulin C-peptide bound to HLA-DQ8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
3SQL
 
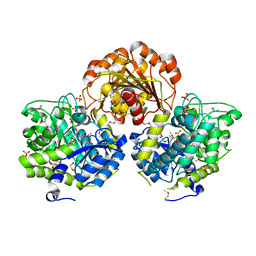 | | Crystal Structure of Glycoside Hydrolase from Synechococcus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase from Synechococcus
To be Published
|
|
4LZB
 
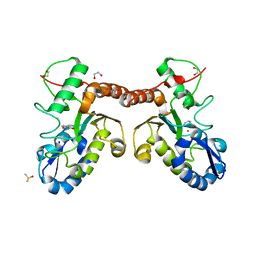 | | Uracil binding pocket in Vaccinia virus uracil DNA glycosylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the uracil complex of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
5IC1
 
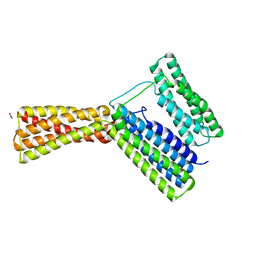 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
