2G1U
 
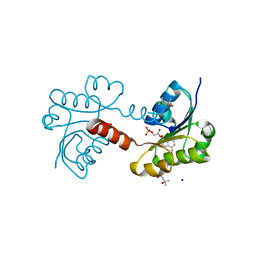 | |
2G8X
 
 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
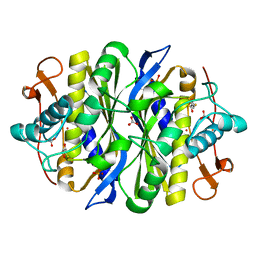
1SJM
 
 | | Nitrite bound copper containing nitrite reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Tocheva, E.I, Rosell, F.I, Mauk, A.G, Murphy, M.E.P. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side-on copper-nitrosyl coordination by nitrite reductase.
Science, 304, 2004
|
|
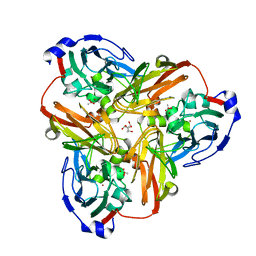
1FDO
 
 | | OXIDIZED FORM OF FORMATE DEHYDROGENASE H FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FORMATE DEHYDROGENASE H, IRON/SULFUR CLUSTER, ... | | Authors: | Sun, P.D, Boyington, J.C. | | Deposit date: | 1997-01-27 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster.
Science, 275, 1997
|
|
1FUN
 
 | | SUPEROXIDE DISMUTASE MUTANT WITH LYS 136 REPLACED BY GLU, CYS 6 REPLACED BY ALA AND CYS 111 REPLACED BY SER (K136E, C6A, C111S) | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE, ... | | Authors: | Lo, T.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Computational, pulse-radiolytic, and structural investigations of lysine-136 and its role in the electrostatic triad of human Cu,Zn superoxide dismutase.
Proteins, 29, 1997
|
|
1GEE
 
 | | Crystal structure of glucose dehydrogenase mutant Q252L complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-07 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
2I9H
 
 | |
4B7K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH CONSENSUS ANKYRIN REPEAT DOMAIN-SER PEPTIDE (20-MER) | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-SER, HYPOXIA-INDUCIBLE FACTOR 1-ALPHA INHIBITOR, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, Ge, W, Schofield, C.J. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate selectivity analyses of factor inhibiting hypoxia-inducible factor.
Angew. Chem. Int. Ed. Engl., 52, 2013
|
|
1HW8
 
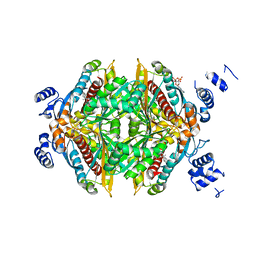 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH COMPACTIN (ALSO KNOWN AS MEVASTATIN) | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,8S,8aR)-2-methyl-8-{[(2S)-2-methylbutanoyl]oxy}-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]h eptanoic acid, ADENOSINE-5'-DIPHOSPHATE, HMG-COA REDUCTASE | | Authors: | Istvan, E.S, Deisenhofer, J. | | Deposit date: | 2001-01-09 | | Release date: | 2001-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural mechanism for statin inhibition of HMG-CoA reductase.
Science, 292, 2001
|
|
1PL3
 
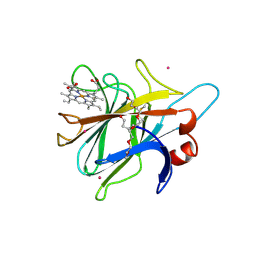 | | Cytochrome Domain Of Cellobiose Dehydrogenase, M65H mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Rotsaert, F.A.J, Hallberg, B.M, de Vries, S, Moenne-Loccoz, P, Divne, C, Gold, M.H. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biophysical and Structural Analysis of a Novel Heme b Iron Ligation in the Flavocytochrome Cellobiose Dehydrogenase.
J.Biol.Chem., 278, 2003
|
|
4ELE
 
 | | Structure-activity relationship guides enantiomeric preference among potent inhibitors of B. anthracis dihydrofolate reductase | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(propan-2-yl)phthalazin-2(1H)-yl]prop-2-en -1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-04-10 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-activity relationship for enantiomers of potent inhibitors of B. anthracis dihydrofolate reductase.
Biochim.Biophys.Acta, 1834, 2013
|
|
3P6P
 
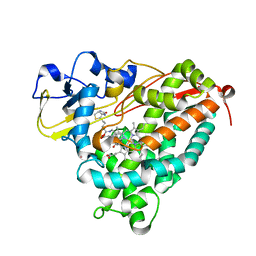 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio | | Descriptor: | (3S,5S,7S)-N-[6-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexyl]tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio
To be Published
|
|
4OEE
 
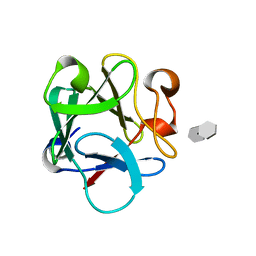 | | Crystal Structure Analysis of FGF2-Disaccharide (S3I2) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
2DV0
 
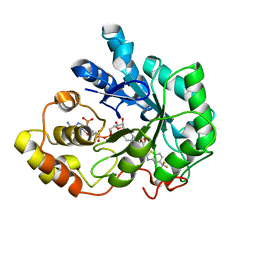 | | Human Aldose Reductase complexed with zopolrestat after 6 days soaking(6days_soaked_2) | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2006-07-28 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions
J.Mol.Biol., 363, 2006
|
|
7EV7
 
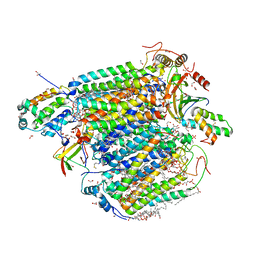 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound fully reduced state at a 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2021-05-20 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of CO-bound fully reduced state of bovine heart cytochrome c oxidase
To Be Published
|
|
1E2R
 
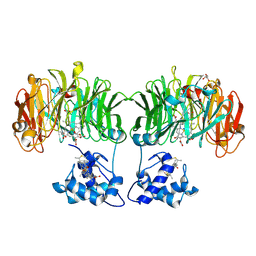 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED AND CYANIDE BOUND | | Descriptor: | CYANIDE ION, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 2000-05-24 | | Release date: | 2000-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-Ray Crystallographic Study of Cyanide Binding Provides Insights Into the Structure-Function Relationship for Cytochrome Cd1 Nitrite Reductase from Paracoccus Pantotrophus.
J.Biol.Chem., 275, 2000
|
|
1DYJ
 
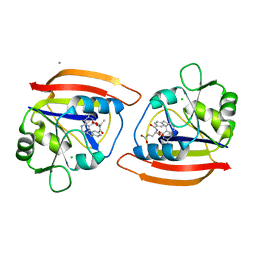 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5,10-DIDEAZATETRAHYDROFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
4OEF
 
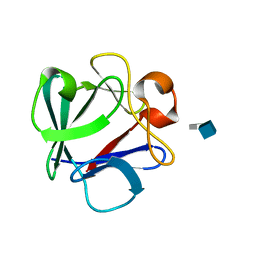 | | Crystal Structure Analysis of FGF2-Disaccharide (S6I2) complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
5QC3
 
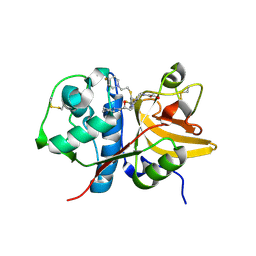 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-{3-[3-{[2-(4-fluoropiperidin-1-yl)ethyl]sulfanyl}-4-(trifluoromethyl)phenyl]-1-[(2S)-2-hydroxy-3-(piperidin-1-yl)propyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}-2-hydroxyethan-1-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
4BDS
 
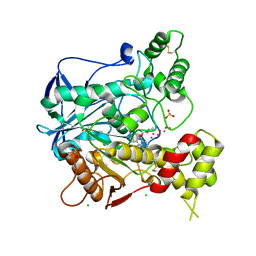 | | Human butyrylcholinesterase in complex with tacrine | | Descriptor: | 1-formyl-L-proline, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Carletti, E, Ronco, C, Trovaslet, M, Nicolet, Y, Jean, L, Renard, P.-Y. | | Deposit date: | 2012-10-06 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Cholinesterases in Complex with Huprine W and Tacrine: Elements of Specificity for Anti-Alzheimer'S Drugs Targeting Acetyl- and Butyrylcholinesterase.
Biochem.J., 453, 2013
|
|
2ZLC
 
 | | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation and crystal structure | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Coactivator peptide DRIP, Vitamin D3 receptor | | Authors: | Shimizu, M, Miyamoto, Y, Nakabayashi, M, Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvitamin D3 analogs: Synthesis, biological evaluation, and crystal structure
Bioorg.Med.Chem., 16, 2008
|
|
4F6W
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 1 (N-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-4-[2-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)ethyl]piperazine-1-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
4OHO
 
 | | Human GKRP bound to AMG-2668 | | Descriptor: | 5-{[(3S)-3-(prop-1-yn-1-yl)-4-{4-[S-(trifluoromethyl)sulfonimidoyl]phenyl}piperazin-1-yl]sulfonyl}pyridin-2-amine, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 3. Structure-activity relationships within the aryl carbinol region of the N-arylsulfonamido-N'-arylpiperazine series.
J.Med.Chem., 57, 2014
|
|
