5CP4
 
 | | CRYOGENIC STRUCTURE OF P450CAM | | Descriptor: | CAMPHOR, CYTOCHROME P450CAM, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 1998-05-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect.
Biochemistry, 37, 1998
|
|
2WCA
 
 | | BtGH84 in complex with n-butyl pugnac | | Descriptor: | CALCIUM ION, O-GLCNACASE BT_4395, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into a Strategy for Attenuating Ampc- Mediated Beta-Lactam Resistance: Structural Basis for Selective Inhibition of the Glycoside Hydrolase Nagz.
Protein Sci., 18, 2009
|
|
4KO3
 
 | | Low X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
6F4D
 
 | |
5L2D
 
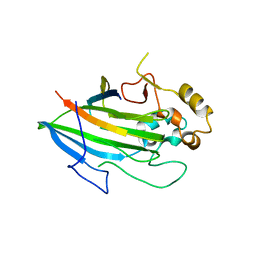 | |
2C3P
 
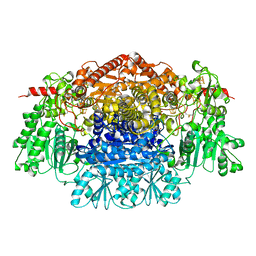 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 1-(2-{(2S,4R,5R)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-[(1S)-1-CARBOXY-1-HYDROXYETHYL]-4-METHYL-1,3-THIAZOLIDIN-5-YL}ETHOXY)-1,1,3,3-TETRAHYDROXY-1LAMBDA~5~-DIPHOSPHOX-1-EN-2-IUM 3-OXIDE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
6FQI
 
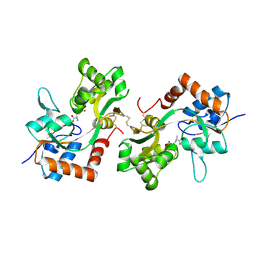 | | GluA2(flop) G724C ligand binding core dimer bound to L-Glutamate (Form B) at 2.91 Angstrom resolution | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.91001153 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
7Q71
 
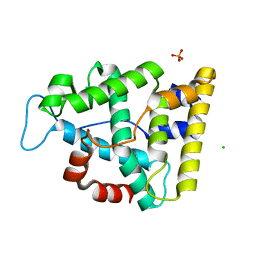 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
2C3M
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
6G94
 
 | |
1DE3
 
 | | SOLUTION STRUCTURE OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | RIBONUCLEASE ALPHA-SARCIN | | Authors: | Perez-Canadillas, J.M, Campos-Olivas, R, Santoro, J, Lacadena, J, Martinez del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 1999-11-12 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity.
J.Mol.Biol., 299, 2000
|
|
2FPQ
 
 | | Crystal Structure of Botulinum Neurotoxin Type D Light Chain | | Descriptor: | BOTULINUM NEUROTOXIN D LIGHT CHAIN, POTASSIUM ION, ZINC ION | | Authors: | Arndt, J.W, Chai, Q, Christian, T, Stevens, R.C. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Botulinum Neurotoxin Type D Light Chain at 1.65 A Resolution: Repercussions for VAMP-2 Substrate Specificity(,).
Biochemistry, 45, 2006
|
|
2FRV
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1997-06-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the [Nife] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands
J.Am.Chem.Soc., 118, 1996
|
|
2C3Y
 
 | | CRYSTAL STRUCTURE OF THE RADICAL FORM OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-13 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
2C3O
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C3U
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus, Oxygen inhibited form | | Descriptor: | 2-(3-{[4-(HYDROXYAMINO)-2-METHYLPYRIMIDIN-5-YL]METHYL}-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL)ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, IRON/SULFUR CLUSTER, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
2C42
 
 | | Crystal Structure Of Pyruvate-Ferredoxin Oxidoreductase From Desulfovibrio africanus | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cavazza, C, Contreras-Martel, C, Pieulle, L, Chabriere, E, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2005-10-14 | | Release date: | 2006-12-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Flexibility of Thiamine Diphosphate Revealed by Kinetic Crystallographic Studies of the Reaction of Pyruvate-Ferredoxin Oxidoreductase with Pyruvate.
Structure, 14, 2006
|
|
6GL4
 
 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | Descriptor: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-22 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
7RQA
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MTI-tripeptidyl-tRNA analog ACCA-ITM at 2.40A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQC
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MFI-tripeptidyl-tRNA analog ACCA-IFM at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6G74
 
 | |
4KN9
 
 | | High-resolution structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KO4
 
 | | High X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
