2MHX
 
 | | Structure of Exocyclic R,R N6,N6-(2,3-Dihydroxy-1,4-butadiyl)-2'-Deoxyadenosine Adduct Induced by 1,2,3,4-Diepoxybutane in DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(RBD)P*AP*GP*AP*AP*G)-3'), 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Kowal, E.A, Seneviratne, U, Wickramaratne, S, Doherty, K.E, Cao, X, Tretyakova, N, Stone, M.P. | | Deposit date: | 2013-12-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Exocyclic R,R- and S,S-N(6),N(6)-(2,3-Dihydroxybutan-1,4-diyl)-2'-Deoxyadenosine Adducts Induced by 1,2,3,4-Diepoxybutane.
Chem.Res.Toxicol., 27, 2014
|
|
2MNX
 
 | | Major groove orientation of the (2S)-N6-(2-hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA adduct induced by 1,2-epoxy-3-butene | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(6HB)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Kowal, E.A, Kotapati, S, Turo, M, Tretyakova, N, Stone, M.P. | | Deposit date: | 2014-04-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Major Groove Orientation of the (2S)-N(6)-(2-Hydroxy-3-buten-1-yl)-2'-deoxyadenosine DNA Adduct Induced by 1,2-Epoxy-3-butene.
Chem.Res.Toxicol., 27, 2014
|
|
213D
 
 | |
1MRE
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRF
 
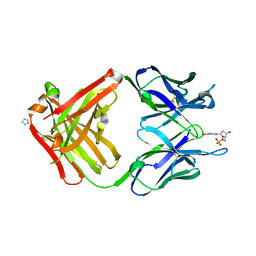 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | 2'-DEOXYINOSINE-5'-MONOPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1MRC
 
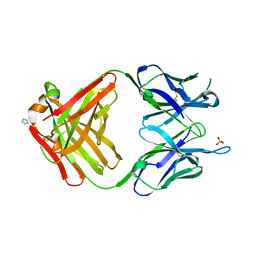 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
214D
 
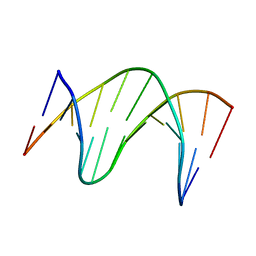 | | THE SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING A SINGLE 2'-O-METHYL-BETA-D-ARAT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*AP*TP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*AP*(T41)P*AP*TP*GP*CP*G)-3') | | Authors: | Gotfredsen, C.H, Spielmann, H.P, Wengel, J, Jacobsen, J.P. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA duplex containing a single 2'-O-methyl-beta-D-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Bioconjug.Chem., 7, 1996
|
|
1HHW
 
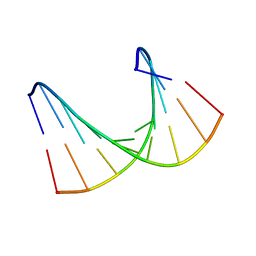 | | Solution structure of LNA1:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1HHX
 
 | | Solution structure of LNA3:RNA hybrid | | Descriptor: | 5- D(*CP*+TP*GP*AP*+TP*AP*+TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1NAO
 
 | | SOLUTION STRUCTURE OF AN RNA 2'-O-METHYLATED RNA DUPLEX CONTAINING AN RNA/DNA HYBRID SEGMENT AT THE CENTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*OMGP*OMUP*OMC)-D(P*AP*TP*CP*T)-R(P*OMCP*OMC)-3'), RNA (5'-R(*GP*GP*AP*GP*AP*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohtsuka, E, Nakamura, H. | | Deposit date: | 1996-03-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA.2'-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center.
Biochemistry, 36, 1997
|
|
1MRD
 
 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), IMIDAZOLE, ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
1OVF
 
 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
1P4Z
 
 | |
227D
 
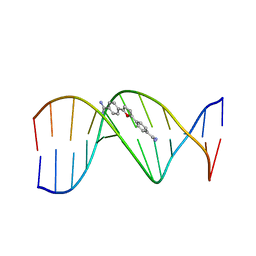 | | A CRYSTALLOGRAPHIC AND SPECTROSCOPIC STUDY OF THE COMPLEX BETWEEN D(CGCGAATTCGCG)2 AND 2,5-BIS(4-GUANYLPHENYL)FURAN, AN ANALOGUE OF BERENIL. STRUCTURAL ORIGINS OF ENHANCED DNA-BINDING AFFINITY | | Descriptor: | 2,5-BIS(4-GUANYLPHENYL)FURAN, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Laughton, C.A, Tanious, F, Nunn, C.M, Boykin, D.W, Wilson, W.D, Neidle, S. | | Deposit date: | 1995-08-08 | | Release date: | 1995-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity.
Biochemistry, 35, 1996
|
|
2C22
 
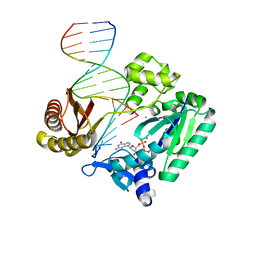 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP*TP *TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
2C2D
 
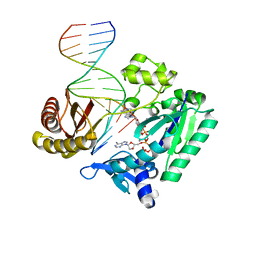 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
2BR0
 
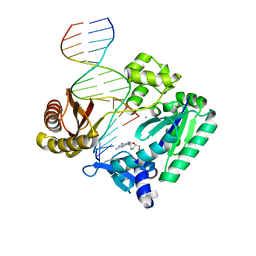 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2C28
 
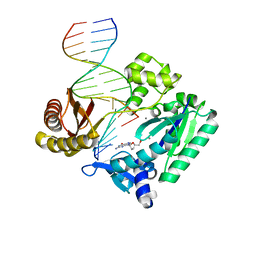 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP*TP *TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
2C2R
 
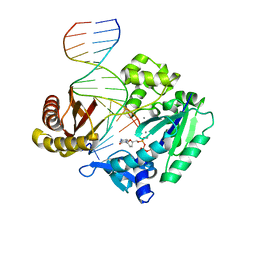 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
2BQU
 
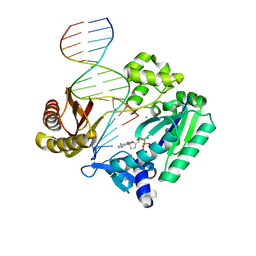 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2C2E
 
 | | Efficient and High Fidelity Incorporation of dCTP Opposite 7,8- Dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA Polymerase Dpo4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*C 8OGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Efficient and High Fidelity Incorporation of Dctp Opposite 7,8-Dihydro-8-Oxodeoxyguanosine by Sulfolobus Solfataricus DNA Polymerase Dpo4
J.Biol.Chem., 281, 2006
|
|
2BQR
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
2BQ3
 
 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', CALCIUM ION, ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
206D
 
 | |
2CCG
 
 | | Crystal structure of His-tagged S. aureus thymidylate kinase complexed with thymidine monophosphate (TMP) | | Descriptor: | THYMIDINE-5'-PHOSPHATE, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
