4UP1
 
 | |
4GN5
 
 | | OBody AM3L15 bound to hen egg-white lysozyme | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Steemson, J.D, Liddament, M.T. | | Deposit date: | 2012-08-16 | | Release date: | 2013-08-21 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
4GN4
 
 | |
4GPM
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR264. | | Descriptor: | Engineered Protein OR264 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4H8F
 
 | |
4HB5
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR267. | | Descriptor: | Engineered Protein | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4HDH
 
 | | Crystal Structure of viral RdRp in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Polyprotein, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
4H8G
 
 | |
4DB9
 
 | | Designed Armadillo repeat protein (YIIIM3AIII) | | Descriptor: | Armadillo repeat protein, YIIIM3AIII | | Authors: | Madhurantakam, C, Varadamsetty, G, Grutter, M.G, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2012-01-13 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based optimization of designed Armadillo-repeat proteins.
Protein Sci., 21, 2012
|
|
4H8O
 
 | |
4H8L
 
 | |
4HDG
 
 | | Crystal Structure of viral RdRp in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Polyprotein, ZINC ION | | Authors: | Surana, P, Nair, D.T. | | Deposit date: | 2012-10-02 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state.
Nucleic Acids Res., 42, 2014
|
|
4DZU
 
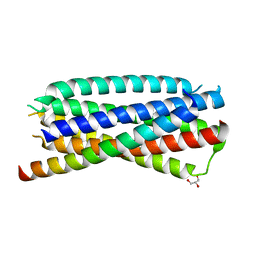 | | Complex of 3-alpha bound to gp41-5 | | Descriptor: | 3-alpha, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
4DZV
 
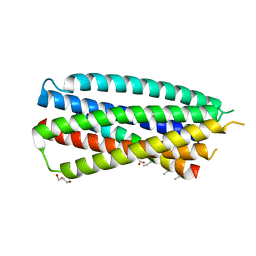 | | Complex of 4-alpha/beta bound to gp41-5 | | Descriptor: | 4-alpha/beta, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
4HQD
 
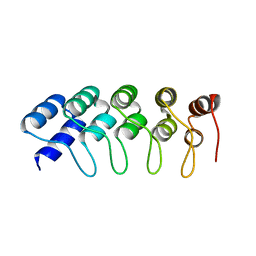 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR265. | | Descriptor: | Engineered Protein OR265 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal Structure of Engineered Protein OR265.
To be Published
|
|
4H8M
 
 | |
4HB1
 
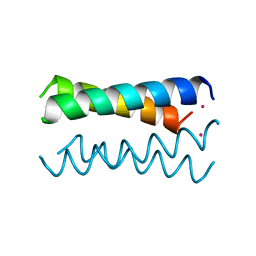 | | A DESIGNED FOUR HELIX BUNDLE PROTEIN. | | Descriptor: | DHP1, UNKNOWN ATOM OR ION | | Authors: | Schafmeister, C.E, Laporte, S.L, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 1997-11-10 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A designed four helix bundle protein with native-like structure.
Nat.Struct.Biol., 4, 1997
|
|
6SS1
 
 | | Kemp Eliminase HG3.17 mutant Q50A, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50A, E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
4OZC
 
 | | Backbone Modifications in the Protein GB1 Helix and Loops: beta-ACPC21, beta-ACPC24, beta-3-Lys28, beta-3-Lys31, beta-ACPC35, beta-ACPC40 | | Descriptor: | GLYCEROL, SULFATE ION, Streptococcal Protein GB1 Backbone Modified Variant: beta-ACPC21, ... | | Authors: | Reinert, Z.E, Horne, W.S. | | Deposit date: | 2014-02-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Folding Thermodynamics of Protein-Like Oligomers with Heterogeneous Backbones.
Chem Sci, 5, 2014
|
|
8SHM
 
 | |
6TU6
 
 | |
8QZN
 
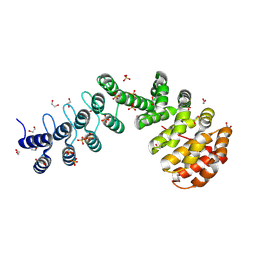 | | DARPin-Armadillo fusion protein with a selected Tyrosin pocket binding to the fused target peptide | | Descriptor: | 1,2-ETHANEDIOL, DARPin-Armadillo fusion protein with Tyrosin pocket and linked target peptide, SULFATE ION | | Authors: | Stark, Y, Menard, F, Jeliazkov, J, Ernst, P, Chembath, A, Ashraf, M, Hine, A, Plueckthun, A. | | Deposit date: | 2023-10-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Modular binder technology by NGS-aided, high-resolution selection in yeast of designed armadillo modules.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4NDJ
 
 | |
8SZZ
 
 | |
