7BDP
 
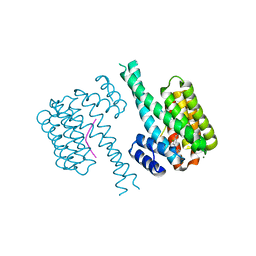 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1017 | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGR
 
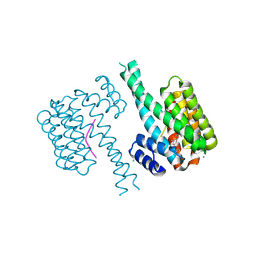 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1016 | | Descriptor: | 14-3-3 protein sigma, 2-methyl-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDT
 
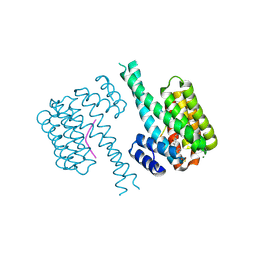 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDY
 
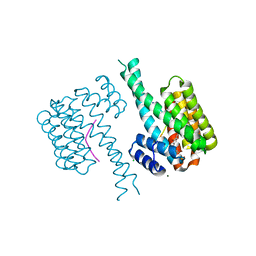 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068B | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-[2-(5-bromanyl-2-fluoranyl-phenyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGV
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1012 | | Descriptor: | 14-3-3 protein sigma, 3-methoxy-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BFW
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068A | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BI3
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-179 | | Descriptor: | 14-3-3 protein sigma, 4-[(6-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)sulfonyl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BG3
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGW
 
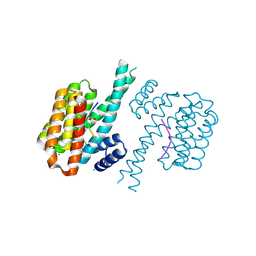 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1011 | | Descriptor: | 14-3-3 protein sigma, 4-(2-phenylimidazol-1-yl)naphthalene-1-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BIY
 
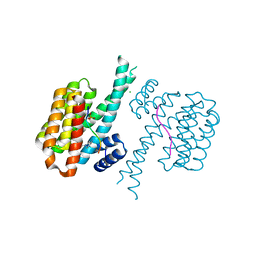 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-175 | | Descriptor: | 1-(4-methanoylphenyl)carbonylpiperidine-4-carbonitrile, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BIW
 
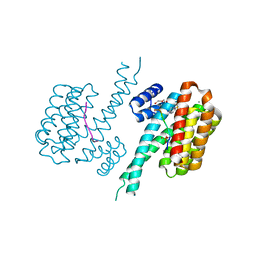 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-187 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinoxalin-1-ylsulfonyl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJB
 
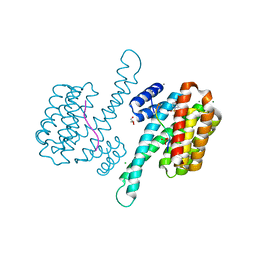 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-044 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methylphenyl)sulfonylmorpholine, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
1PFM
 
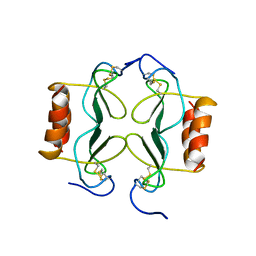 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 1-15 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
1PFN
 
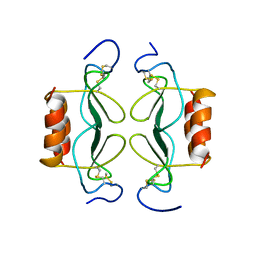 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
4H10
 
 | |
7XBD
 
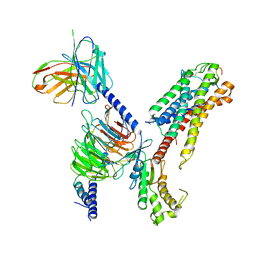 | | Cryo-EM structure of human galanin receptor 2 | | Descriptor: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ishimoto, N, Kita, S, Park, S.Y. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
1P63
 
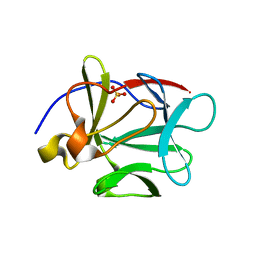 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
2R6M
 
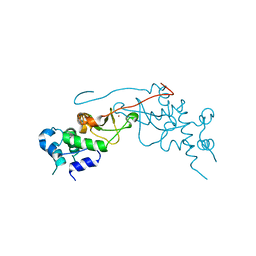 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
6EY7
 
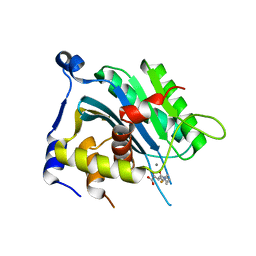 | | Human cytomegalovirus terminase nuclease domain, Mn soaked, inhibitor bound | | Descriptor: | 4-[(4-fluorophenyl)methyl-methyl-amino]-2,4-bis(oxidanylidene)butanoic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bongarzone, S, Nadal, M, Kaczmarska, Z, Machon, C, Alvarez, M, Albericio, F, Coll, M. | | Deposit date: | 2017-11-10 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Driven Discovery of alpha , gamma-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase.
Acs Omega, 3, 2018
|
|
1W22
 
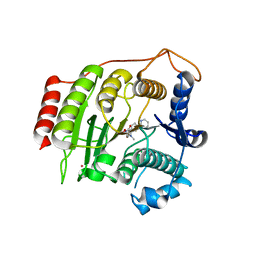 | | Crystal structure of inhibited human HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, N-HYDROXY-4-(METHYL{[5-(2-PYRIDINYL)-2-THIENYL]SULFONYL}AMINO)BENZAMIDE, POTASSIUM ION, ... | | Authors: | Vannini, A, Volpari, C, Caroli Casavola, E, Di Marco, S. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Eukaryotic Zn-Dependent Histone Deacetylase,Human Hdac8,Complexed with a Hydroxamic Acid Inhibitor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
7AW9
 
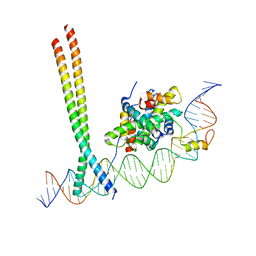 | | CCAAT-binding complex and HapX bound to Aspergillus fumigatus cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
7AW7
 
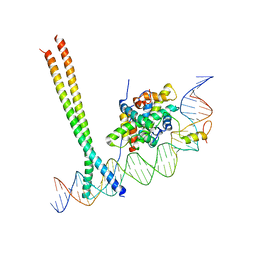 | | CCAAT-binding complex and HapX bound to Aspergillus nidulans cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
2AYU
 
 | |
3TVD
 
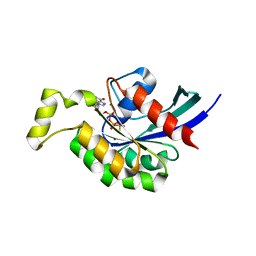 | | Crystal Structure of Mouse RhoA-GTP complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Swaminathan, K, Pal, K, Jobichen, C. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Crystal structure of mouse RhoA:GTPgammaS complex in a centered lattice.
J.Struct.Funct.Genom., 13, 2012
|
|
1QP9
 
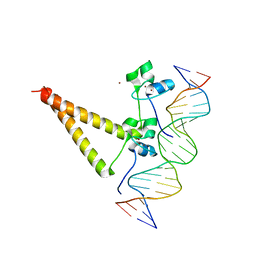 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
