3DRG
 
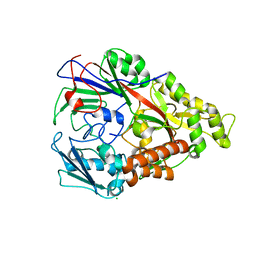 | | Lactococcal OppA complexed with bradykinin in the closed conformation | | Descriptor: | Bradykinin, CHLORIDE ION, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DRK
 
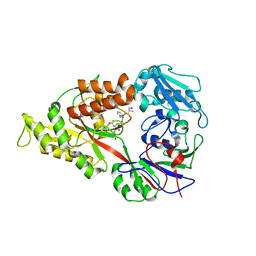 | | Crystal structure of Lactococcal OppA co-crystallized with Neuropeptide S in an open conformation | | Descriptor: | Neuropeptide S, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
2QUQ
 
 | |
2R2O
 
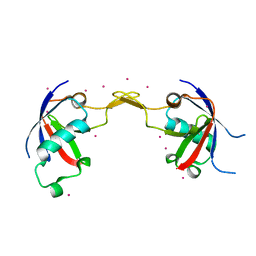 | | Crystal structure of the effector domain of human Plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION | | Authors: | Tong, Y, Tempel, W, Shen, L, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-27 | | Release date: | 2007-09-04 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain.
J.Biol.Chem., 282, 2007
|
|
2QW9
 
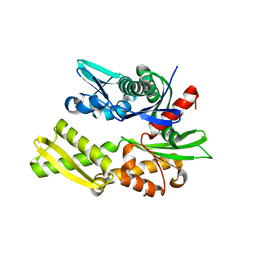 | | Crystal structure of bovine hsc70 (1-394aa)in the apo state | | Descriptor: | GLYCEROL, Heat shock cognate 71 kDa protein | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
2QXI
 
 | | High resolution structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3DP3
 
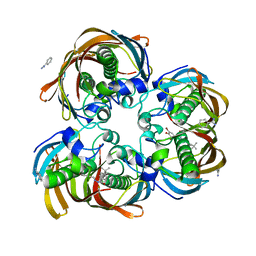 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
2PR8
 
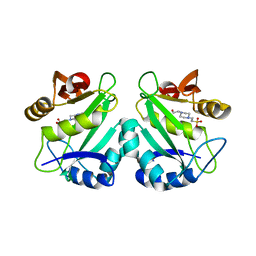 | | crystal structure of aminoglycoside N-acetyltransferase AAC(6')-Ib11 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside 6-N-acetyltransferase type Ib11 | | Authors: | Maurice, F, Broutin, I, Podglajen, I, Benas, P, Collatz, E, Dardel, F. | | Deposit date: | 2007-05-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance.
Embo Rep., 9, 2008
|
|
2PRV
 
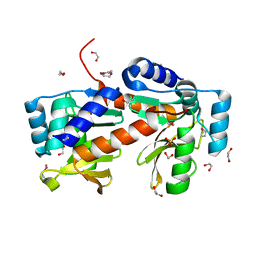 | |
2PV4
 
 | |
2Q82
 
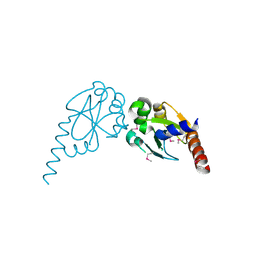 | | Crystal structure of core protein P7 from Pseudomonas phage phi12. Northeast Structural Genomics Target OC1 | | Descriptor: | Core protein P7 | | Authors: | Benach, J, Eryilmaz, E, Su, M, Seetharaman, J, Wei, H, Gottlieb, P, Hunt, J.F, Ghose, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-08 | | Release date: | 2007-08-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and dynamics of the P7 protein from the bacteriophage phi 12.
J.Mol.Biol., 382, 2008
|
|
3BMO
 
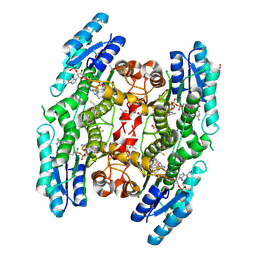 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX4) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-[(4-methylphenyl)sulfanyl]pyrimidine-2,4-diamine, ... | | Authors: | Martini, V.P, Iulek, J, Hunter, W.N, Tulloch, L.B. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3BNL
 
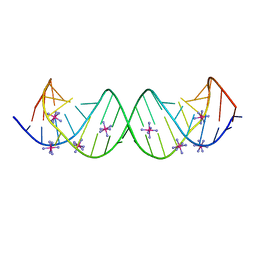 | |
3CAI
 
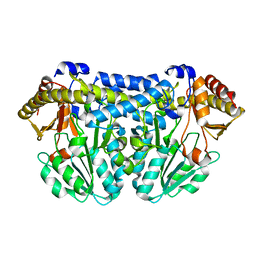 | | Crystal structure of Mycobacterium tuberculosis Rv3778c protein | | Descriptor: | POSSIBLE AMINOTRANSFERASE | | Authors: | Covarrubias, A.S, Larsson, A.M, Jones, T.A, Bergfors, T, Mowbray, S.L, Unge, T. | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical function studies of Mycobacterium tuberculosis essential protein Rv3778c
To be published
|
|
3BXX
 
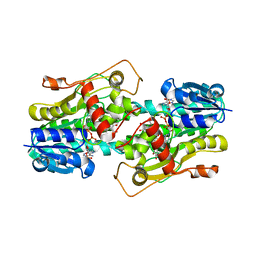 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-15 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3BRX
 
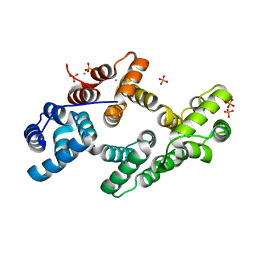 | | Crystal Structure of calcium-bound cotton annexin Gh1 | | Descriptor: | Annexin, CALCIUM ION, PHOSPHATE ION | | Authors: | Hu, N.-J, Hofmann, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-bound annexin Gh1 from Gossypium hirsutum and its implications for membrane binding mechanisms of plant annexins.
J.Biol.Chem., 283, 2008
|
|
3CES
 
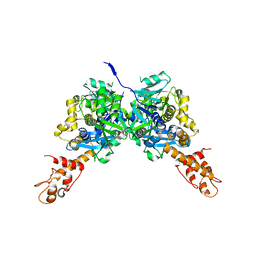 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3COQ
 
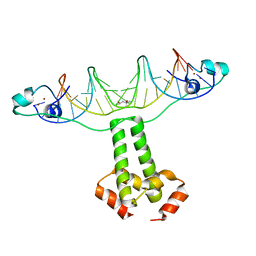 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
3CVE
 
 | | Crystal Structure of the carboxy terminus of Homer1 | | Descriptor: | Homer protein homolog 1 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
3D27
 
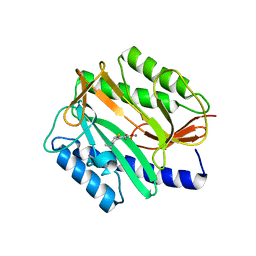 | | E. coli methionine aminopeptidase with Fe inhibitor W29 | | Descriptor: | 4-(3-ethylthiophen-2-yl)benzene-1,2-diol, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Chai, S, He, H.Z. | | Deposit date: | 2008-05-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of inhibitors of Escherichia coli methionine aminopeptidase with the Fe(II)-form selectivity and antibacterial activity.
J.Med.Chem., 51, 2008
|
|
3D32
 
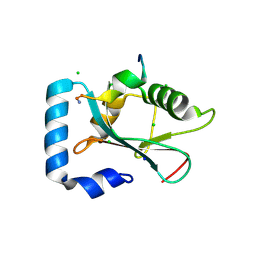 | | Complex of GABA(A) receptor-associated protein (GABARAP) with a synthetic peptide | | Descriptor: | CHLORIDE ION, Gamma-aminobutyric acid receptor-associated protein, K1 peptide, ... | | Authors: | Weiergraeber, O.H, Stangler, T, Willbold, D. | | Deposit date: | 2008-05-09 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Binding Mode of GABA(A) Receptor-Associated Protein.
J.Mol.Biol., 381, 2008
|
|
2OA2
 
 | |
3CFM
 
 | |
3CIN
 
 | |
2OKC
 
 | |
