4I4V
 
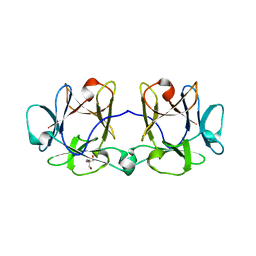 | | BEL beta-trefoil complex with N-acetylgalactosamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, BEL beta-trefoil | | Authors: | Bovi, M, Cenci, L, Perduca, M, Capaldi, S, Carrizo, M.E, Civiero, L, Chiarelli, L.R, Galliano, M, Monaco, H.L. | | Deposit date: | 2012-11-28 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | BEL {beta}-trefoil: A novel lectin with antineoplastic properties in king bolete (Boletus edulis) mushrooms.
Glycobiology, 23, 2013
|
|
3TS7
 
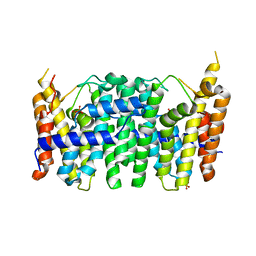 | | CRYSTAL STRUCTURE OF FARNESYL DIPHOSPHATE SYNTHASE (TARGET EFI-501951) FROM Methylococcus capsulatus | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IJG
 
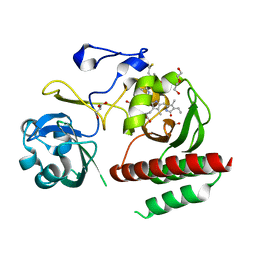 | | Crystal structure of monomeric bacteriophytochrome | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Auldridge, M.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
8TG8
 
 | |
1ILV
 
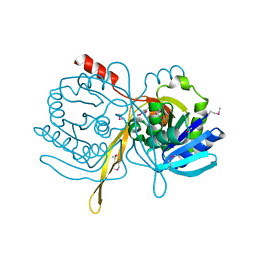 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
4A23
 
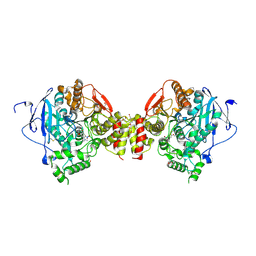 | | Mus musculus Acetylcholinesterase in complex with racemic C5685 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Andersson, C.D, Artrursson, E, Hornberg, A, Tunemalm, A.K, Linusson, A, Ekstrom, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling.
Plos One, 6, 2011
|
|
1M7G
 
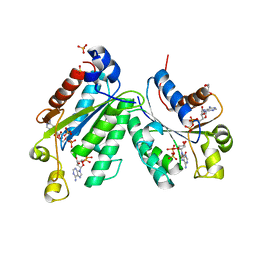 | | Crystal structure of APS kinase from Penicillium Chrysogenum: Ternary structure with ADP and APS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-2',3'-VANADATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Lansdon, E.B, Segel, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
1EHK
 
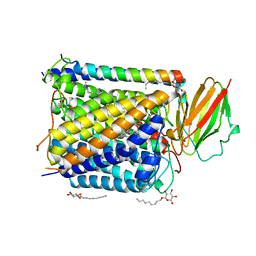 | | CRYSTAL STRUCTURE OF THE ABERRANT BA3-CYTOCHROME-C OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | BA3-TYPE CYTOCHROME-C OXIDASE, COPPER (II) ION, DINUCLEAR COPPER ION, ... | | Authors: | Soulimane, T, Buse, G, Bourenkov, G.P, Bartunik, H.D, Huber, R, Than, M.E. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus.
EMBO J., 19, 2000
|
|
3SZE
 
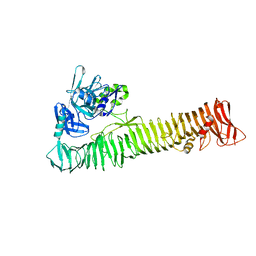 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
4F04
 
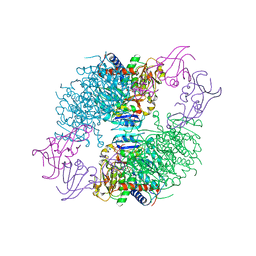 | | A Second Allosteric site in E. coli Aspartate Transcarbamoylase: R-state ATCase with UTP bound | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Peterson, A.W, Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A second allosteric site in Escherichia coli aspartate transcarbamoylase.
Biochemistry, 51, 2012
|
|
2FCP
 
 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-15 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
2R8A
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta N8 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From the Cover: Ligand-gated diffusion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4FGU
 
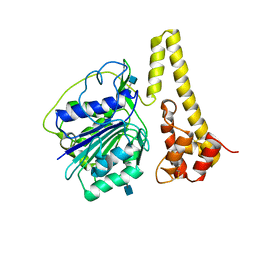 | | Crystal structure of prolegumain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain | | Authors: | Dall, E, Brandstetter, H. | | Deposit date: | 2012-06-05 | | Release date: | 2013-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2FTS
 
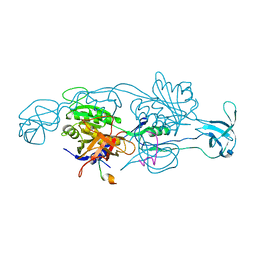 | |
1EPA
 
 | |
2FPU
 
 | | Crystal Structure of the N-terminal domain of E.coli HisB- Complex with histidinol | | Descriptor: | CHLORIDE ION, Histidine biosynthesis bifunctional protein hisB, L-histidinol, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
4FZW
 
 | | Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex from E.coli | | Descriptor: | 1,2-epoxyphenylacetyl-CoA isomerase, 2,3-dehydroadipyl-CoA hydratase, GLYCEROL | | Authors: | Grishin, A.M, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-07-08 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Protein Interactions in the beta-Oxidation Part of the Phenylacetate
Utilization Pathway. Crystal Structure of the PaaF-PaaG Hydratase-Isomerase Complex
J.Biol.Chem., 287, 2012
|
|
4FXP
 
 | |
3P7I
 
 | |
1LMQ
 
 | |
2FFX
 
 | | Structure of Human Ferritin L. Chain | | Descriptor: | CADMIUM ION, SULFATE ION, ferritin light chain | | Authors: | Wang, Z.M, Li, C, Ellenburg, M.P, Soitsman, E.M, Ruble, J.R, Wright, B.S, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human ferritin L chain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1DQI
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE FROM P. FURIOSUS IN THE OXIDIZED STATE AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
2Y42
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with NADH and Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, BICINE, MANGANESE (II) ION, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
1DUV
 
 | | CRYSTAL STRUCTURE OF E. COLI ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH NDELTA-L-ORNITHINE-DIAMINOPHOSPHINYL-N-SULPHONIC ACID (PSORN) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDELTA-(N'-SULPHODIAMINOPHOSPHINYL)-L-ORNITHINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Langley, D.B, Templeton, M.D, Fields, B.A, Mitchell, R.E, Collyer, C.A. | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N'-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism.
J.Biol.Chem., 275, 2000
|
|
3SB2
 
 | | Crystal Structure of the RNA chaperone Hfq from Herbaspirillum seropedicae SMR1 | | Descriptor: | GLYCEROL, Protein hfq | | Authors: | Kadowaki, M.A.S, Iulek, J, Barbosa, J.A.R.G, Pedrosa, F.O, Souza, E.M, Chubatsu, L.S, Monteiro, R.A, Steffens, M.B.R. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6301 Å) | | Cite: | Structural characterization of the RNA chaperone Hfq from the nitrogen-fixing bacterium Herbaspirillum seropedicae SmR1.
Biochim.Biophys.Acta, 1824, 2011
|
|
