5IRI
 
 | |
6RA6
 
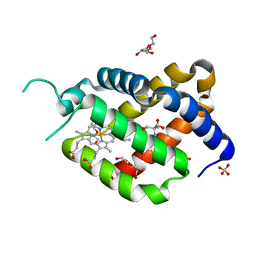 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
8WGR
 
 | | Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with antidepressant desipramine in KCl condition. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
7ZDB
 
 | |
7ZDG
 
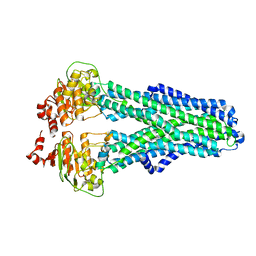 | | IF(heme/confined) conformation of CydDC (Dataset-5) | | Descriptor: | ATP-binding/permease protein CydC, ATP-binding/permease protein CydD, HEME B/C | | Authors: | Wu, D, Safarian, S. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Dissecting the conformational complexity and mechanism of a bacterial heme transporter.
Nat.Chem.Biol., 19, 2023
|
|
7ZDL
 
 | |
7ZEC
 
 | |
6TOL
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound 25a | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-[(2~{S},6~{R})-2,6-dimethylmorpholin-4-yl]pyrimidin-4-yl]amino]-1-methyl-3-(3-methyl-3-oxidanyl-butyl)benzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
6TOS
 
 | | Crystal structure of the Orexin-1 receptor in complex with GSK1059865 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6G6W
 
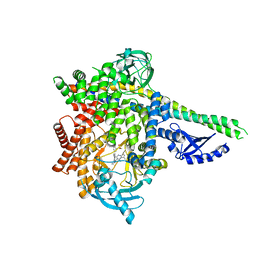 | | HUMAN PI3KDELTA IN COMPLEX WITH LIGAND LASW1976 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[3-[4-[[(1~{S})-1-(5-methyl-4-oxidanylidene-3-phenyl-pyrrolo[2,1-f][1,2,4]triazin-2-yl)ethyl]amino]-7~{H}-pyrrolo[ 2,3-d]pyrimidin-5-yl]-5-oxidanyl-phenyl]methanesulfonamide | | Authors: | Segarra, V, Hernandez, B, Jestel, A, Mortel, M, Nagel, S. | | Deposit date: | 2018-04-03 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery of a Novel Inhaled PI3K delta Inhibitor for the Treatment of Respiratory Diseases.
J. Med. Chem., 61, 2018
|
|
8JCC
 
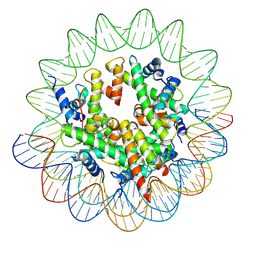 | |
5V3B
 
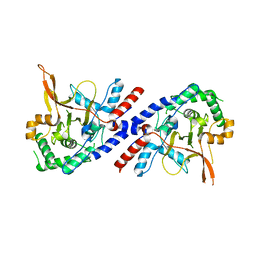 | |
7LF4
 
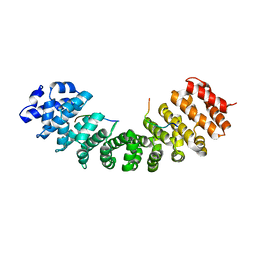 | |
5VN8
 
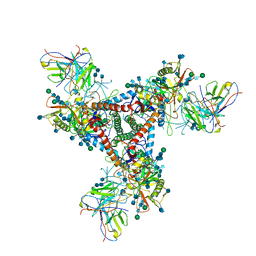 | | Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Pallesen, J, Ward, A.B, Cottrell, C.A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
7TUK
 
 | |
5JG6
 
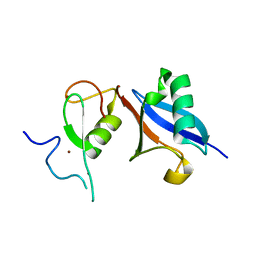 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
6TOJ
 
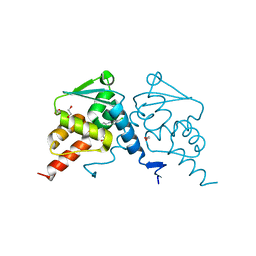 | | Crystal structure of human BCL6 BTB domain in complex with compound 17a | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[1-methyl-3-(3-methyl-3-oxidanyl-butyl)-2-oxidanylidene-benzimidazol-5-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein | | Authors: | Rodrigues, M.J, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | AchievingIn VivoTarget Depletion through the Discovery and Optimization of Benzimidazolone BCL6 Degraders.
J.Med.Chem., 63, 2020
|
|
6WDC
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure IV-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WDE
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
5VBN
 
 | | Crystal Structure of human DNA polymerase epsilon B-subunit in complex with C-terminal domain of catalytic subunit | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit 2, SULFATE ION, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2017-03-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human Pol B-subunit in complex with the C-terminal domain of the catalytic subunit.
J. Biol. Chem., 292, 2017
|
|
1TMN
 
 | |
4WSA
 
 | | Crystal structure of Influenza B polymerase bound to the vRNA promoter (FluB1 form) | | Descriptor: | Influenza B vRNA promoter 3' end, Influenza B vRNA promoter 5' end, PA, ... | | Authors: | Reich, S, Guilligay, D, Pflug, A, Cusack, S. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into cap-snatching and RNA synthesis by influenza polymerase.
Nature, 516, 2014
|
|
6WDG
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
3P2P
 
 | | ENHANCED ACTIVITY AND ALTERED SPECIFICITY OF PHOSPHOLIPASE A2 BY DELETION OF A SURFACE LOOP | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Thunnissen, M.M.G.M, Kalk, K.H, Drenth, J. | | Deposit date: | 1989-11-29 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced activity and altered specificity of phospholipase A2 by deletion of a surface loop.
Science, 244, 1989
|
|
