2GCG
 
 | | Ternary Crystal Structure of Human Glyoxylate Reductase/Hydroxypyruvate Reductase | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, Glyoxylate reductase/hydroxypyruvate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Booth, M.P.S, Conners, R, Rumsby, G, Brady, R.L. | | Deposit date: | 2006-03-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate specificity in human glyoxylate reductase/hydroxypyruvate reductase
J.Mol.Biol., 360, 2006
|
|
2GCH
 
 | |
2GCI
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GCJ
 
 | | Crystal Structure of the Pob3 middle domain | | Descriptor: | Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2GCL
 
 | | Structure of the Pob3 Middle domain | | Descriptor: | CHLORIDE ION, Hypothetical 63.0 kDa protein in DAK1-ORC1 intergenic region | | Authors: | VanDemark, A.P. | | Deposit date: | 2006-03-14 | | Release date: | 2006-05-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Structure of the yFACT Pob3-M Domain, Its Interaction with the DNA Replication Factor RPA, and a Potential Role in Nucleosome Deposition.
Mol.Cell, 22, 2006
|
|
2GCN
 
 | |
2GCO
 
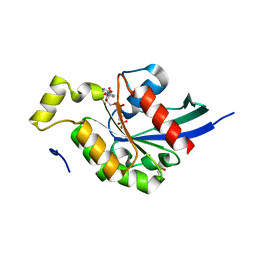 | | Crystal structure of the human RhoC-GppNHp complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rho-related GTP-binding protein RhoC | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
2GCP
 
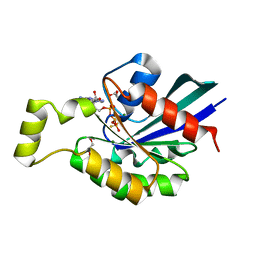 | | Crystal structure of the human RhoC-GSP complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dias, S.M.G, Cerione, R.A. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Crystal Structures Reveal Two Activated States for RhoC.
Biochemistry, 46, 2007
|
|
2GCQ
 
 | |
2GCS
 
 | |
2GCT
 
 | |
2GCU
 
 | | X-Ray Structure of Gene Product from Arabidopsis Thaliana At1g53580 | | Descriptor: | 1,2-ETHANEDIOL, FE (II) ION, Putative hydroxyacylglutathione hydrolase 3, ... | | Authors: | McCoy, J.G, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-14 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Structure of an ETHE1-like protein from Arabidopsis thaliana.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2GCV
 
 | |
2GCX
 
 | | Solution Structure of Ferrous Iron Transport Protein A (FeoA) of Klebsiella pneumoniae | | Descriptor: | Ferrous iron transport protein A | | Authors: | Hung, K.-W, Cheng, C.-C, Yu, T.-H, Wang, S.-H, Chang, C.-F, Tsai, S.-F, Huang, T.-H. | | Deposit date: | 2006-03-14 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ferrous Iron Transport Protein A (FeoA) of Klebsiella pneumoniae
To be Published
|
|
2GCY
 
 | |
2GCZ
 
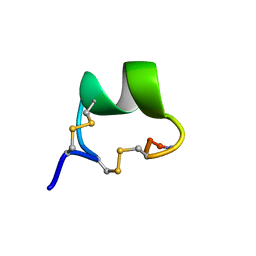 | | Solution Structure of alpha-Conotoxin OmIA | | Descriptor: | Alpha-conotoxin OmIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-03-15 | | Release date: | 2006-07-25 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist alpha-conotoxin OmIA that discriminates alpha3 vs. alpha6 nAChR subtypes
Biochem.Biophys.Res.Commun., 345, 2006
|
|
2GD0
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (S)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD1
 
 | |
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD3
 
 | | NMR structure of S14G-humanin in 30% TFE solution | | Descriptor: | Humanin | | Authors: | Benaki, D, Zikos, C, Evangelou, A, Livaniou, E, Vlassi, M, Mikros, E, Pelecanou, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ser14Gly-humanin, a potent rescue factor against neuronal cell death in Alzheimer's disease.
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2GD4
 
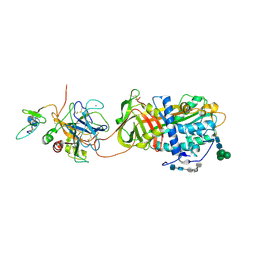 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
2GD5
 
 | | Structural basis for budding by the ESCRTIII factor CHMP3 | | Descriptor: | Charged multivesicular body protein 3 | | Authors: | Muziol, T.M, Pineda-Molina, E, Ravelli, R.B, Zamborlini, A, Usami, Y, Gottlinger, H, Weissenhorn, W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Budding by the ESCRT-III Factor CHMP3.
Dev.Cell, 10, 2006
|
|
2GD6
 
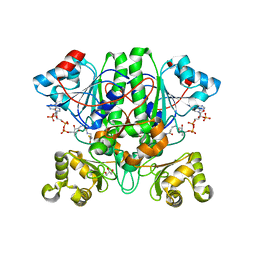 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD7
 
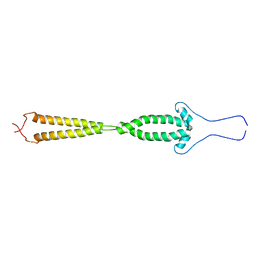 | |
2GD8
 
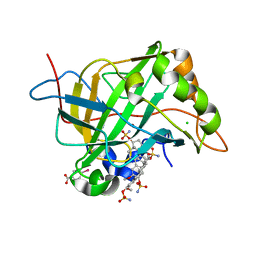 | | Crystal structure analysis of the human carbonic anhydrase II in complex with a 2-substituted estradiol bis-sulfamate | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-METHOXYESTRA-1,3,5(10)-TRIENE-3,17-DIYL DISULFAMATE, CHLORIDE ION, Carbonic anhydrase 2, ... | | Authors: | De Simone, G, Di Fiore, A. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | 2-substituted estradiol bis-sulfamates, multitargeted antitumor agents: synthesis, in vitro SAR, protein crystallography, and in vivo activity.
J.Med.Chem., 49, 2006
|
|
