2QXJ
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone and Copper | | Descriptor: | COPPER (II) ION, Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2RHP
 
 | | The Thrombospondin-1 Polymorphism Asn700Ser Associated with Cornoary Artery Disease Causes Local and Long-Ranging Changes in Protein Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Carlson, C.B, Keck, J.L, Mosher, D.F. | | Deposit date: | 2007-10-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Influences of the N700S Thrombospondin-1 Polymorphism on Protein Structure and Stability.
J.Biol.Chem., 283, 2008
|
|
1YXP
 
 | | HIV-1 DIS RNA subtype F- Zn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', ZINC ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-02-22 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
1XPE
 
 | | HIV-1 subtype B genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XP7
 
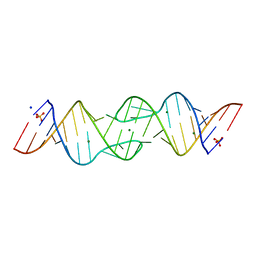 | | HIV-1 subtype F genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XPF
 
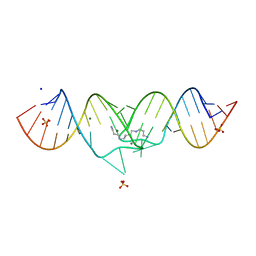 | | HIV-1 subtype A genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1ZCI
 
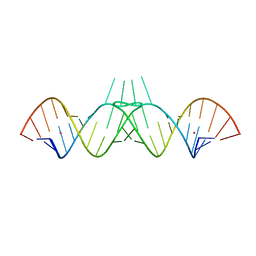 | | HIV-1 DIS RNA subtype F- monoclinic form | | Descriptor: | 5'-R(*CP*(5BU)P*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
1Y3S
 
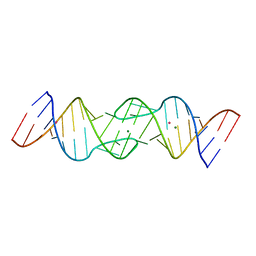 | | HIV-1 DIS RNA subtype F- MPD form | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, POTASSIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
1Y3O
 
 | | HIV-1 DIS RNA subtype F- Mn soaked | | Descriptor: | 5'-R(*CP*UP*(5BU)P*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, SODIUM ION | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-11-26 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies.
J.Mol.Biol., 356, 2006
|
|
2M99
 
 | |
1FON
 
 | | CRYSTAL STRUCTURE OF BOVINE PROCARBOXYPEPTIDASE A-S6 SUBUNIT III, A HIGHLY STRUCTURED TRUNCATED ZYMOGEN E | | Descriptor: | PROCARBOXYPEPTIDASE A-S6 | | Authors: | Pignol, D.C, Gaboriaud, T, Michon, B, Kerfelec, B, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of bovine procarboxypeptidase A-S6 subunit III, a highly structured truncated zymogen E.
EMBO J., 13, 1994
|
|
1EKM
 
 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
3LOY
 
 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
1HRN
 
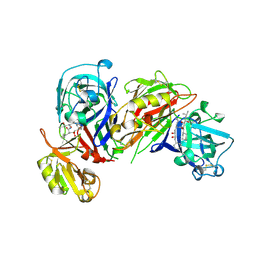 | | HIGH RESOLUTION CRYSTAL STRUCTURES OF RECOMBINANT HUMAN RENIN IN COMPLEX WITH POLYHYDROXYMONOAMIDE INHIBITORS | | Descriptor: | (2R,4S,5S)-N-[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]-2-(cyclopropylmethyl)-4,5-dihydroxy-6-phenylhexanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, RENIN | | Authors: | Tong, L, Anderson, P.C. | | Deposit date: | 1995-03-31 | | Release date: | 1995-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution crystal structures of recombinant human renin in complex with polyhydroxymonoamide inhibitors.
J.Mol.Biol., 250, 1995
|
|
3E4C
 
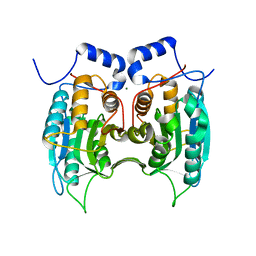 | | Procaspase-1 zymogen domain crystal structure | | Descriptor: | Caspase-1, MAGNESIUM ION | | Authors: | Elliott, J.M, Rouge, L, Wiesmann, C, Scheer, J.M. | | Deposit date: | 2008-08-11 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of procaspase-1 zymogen domain reveals insight into inflammatory caspase autoactivation
J.Biol.Chem., 284, 2009
|
|
1EVJ
 
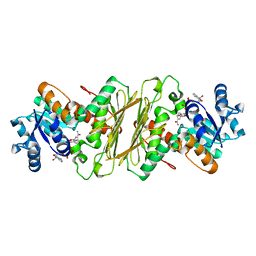 | | CRYSTAL STRUCTURE OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE (GFOR) DELTA1-22 S64D | | Descriptor: | GLUCOSE-FRUCTOSE OXIDOREDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lott, J.S, Halbig, D, Baker, H.M, Hardman, M.J, Sprenger, G.A, Baker, E.N. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation.
J.Mol.Biol., 304, 2000
|
|
1HF4
 
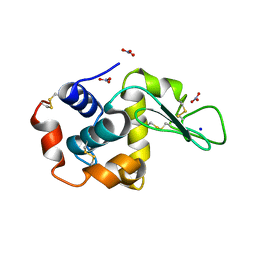 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3I3Q
 
 | | Crystal Structure of AlkB in complex with Mn(II) and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB, MANGANESE (II) ION | | Authors: | Yu, B, Hunt, J.F. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzymological and structural studies of the mechanism of promiscuous substrate recognition by the oxidative DNA repair enzyme AlkB.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3I2O
 
 | |
3I3M
 
 | |
3I49
 
 | |
2MN2
 
 | | 3D structure of YmoB, a modulator of biofilm formation | | Descriptor: | YmoB | | Authors: | Marimon, O, Cordeiro, T.N, Amata, I, Pons, M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An oxygen-sensitive toxin-antitoxin system.
Nat Commun, 7, 2016
|
|
1GHA
 
 | | A SECOND ACTIVE SITE IN CHYMOTRYPSIN? THE X-RAY CRYSTAL STRUCTURE OF N-ACETYL-D-TRYPTOPHAN BOUND TO GAMMA-CHYMOTRYPSIN | | Descriptor: | GAMMA-CHYMOTRYPSIN A, ISOPROPYL ALCOHOL, PRO GLY VAL TYR PEPTIDE, ... | | Authors: | Yennawar, H.P, Yennawar, N.H, Farber, G.K. | | Deposit date: | 1994-04-06 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A STRUCTURAL EXPLANATION FOR ENZYME MEMORY IN NONAQUEOUS SOLVENTS.
J.Am.Chem.Soc., 117, 1995
|
|
7O3I
 
 | | Cystal structure of Zymogen Granule Protein 16 (ZG16) | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational switches and redox properties of the colon cancer-associated human lectin ZG16.
Febs J., 288, 2021
|
|
7O4P
 
 | | Cystal structure of Zymogen Granule Protein 16 (ZG16) | | Descriptor: | CHLORIDE ION, GLYCEROL, Zymogen granule membrane protein 16 | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-04-07 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Conformational switches and redox properties of the colon cancer-associated human lectin ZG16.
Febs J., 288, 2021
|
|
