1H9R
 
 | | Tungstate bound complex Dimop domain of ModE from E.coli | | Descriptor: | MOLYBDENUM TRANSPORT PROTEIN MODE, NICKEL (II) ION, TUNGSTATE(VI)ION | | Authors: | Gourley, D.G, Hunter, W.N. | | Deposit date: | 2001-03-19 | | Release date: | 2001-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxyanion Binding Alters Conformational and Quaternary Structure of the C-Terminal Domain of the Transcriptional Regulator Mode; Implications for Molybdate-Dependant Regulation, Signalling, Storage and Transport
J.Biol.Chem., 276, 2001
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
6DGB
 
 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
5CXH
 
 | |
4YYX
 
 | |
3AG3
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ABL
 
 | | Bovine heart cytochrome c oxidase at the fully oxidized state (15-s X-ray exposure dataset) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Aoyama, H, Muramoto, K, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Ogura, T, Yoshikawa, S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6OD2
 
 | | Yeast Spc42 C-terminal Antiparallel Coiled-Coil | | Descriptor: | Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
7MKI
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6EQ9
 
 | | Crystal structure of JNK3 in complex with AMP-PCP | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Macedo, J.T, Stehle, T, Blaum, B.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Optimization of a Pyridinylimidazole Scaffold: Shifting the Selectivity from p38 alpha Mitogen-Activated Protein Kinase to c-Jun N-Terminal Kinase 3.
ACS Omega, 3, 2018
|
|
1P7N
 
 | | Dimeric Rous Sarcoma virus Capsid protein structure with an upstream 25-amino acid residue extension of C-terminal of Gag p10 protein | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Nandhagopal, N, Simpson, A.A, Johnson, M.C, Francisco, A.B, Schatz, G.W, Rossmann, M.G, Vogt, V.M. | | Deposit date: | 2003-05-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimeric rous sarcoma virus capsid protein structure relevant to immature gag assembly
J.Mol.Biol., 335, 2004
|
|
7CGL
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGI
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGK
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGF
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGG
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGH
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGM
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGJ
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
6UE3
 
 | |
6UL3
 
 | |
1HSQ
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
5B16
 
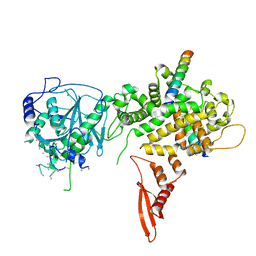 | | X-ray structure of DROSHA in complex with the C-terminal tail of DGCR8. | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3,DROSHA,Ribonuclease 3,DROSHA,Ribonuclease 3, ZINC ION | | Authors: | Kwon, S.C, Nguyen, T.A, Choi, Y.G, Jo, M.H, Hohng, S, Kim, V.N, Woo, J.S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human DROSHA
Cell, 164, 2016
|
|
2WXT
 
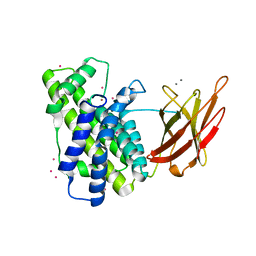 | | Clostridium perfringens alpha-toxin strain NCTC8237 | | Descriptor: | CADMIUM ION, CALCIUM ION, PHOSPHOLIPASE C, ... | | Authors: | Justin, N, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WXU
 
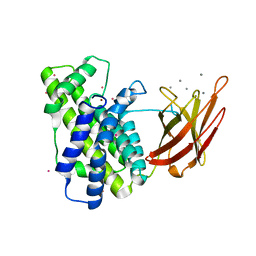 | | Clostridium perfringens alpha-toxin strain NCTC8237 mutant T74I | | Descriptor: | CADMIUM ION, CALCIUM ION, PHOSPHOLIPASE C, ... | | Authors: | Vachieri, S.G, Naylor, C.E, Basak, A.K. | | Deposit date: | 2009-11-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of a Nontoxic Variant of Clostridium Perfringens [Alpha]-Toxin with the Toxic Wild-Type Strain
Acta Crystallogr.,Sect.D, 66, 2010
|
|
