5MTJ
 
 | | Yes1-SH2 in complex with monobody Mb(Yes_1) | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Monobody Mb(Yes_1), SULFATE ION, ... | | Authors: | Sha, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
5N1O
 
 | |
8TUF
 
 | |
8UEA
 
 | | Crystal structure of SARS-CoV-2 3CL protease with inhibitor 29 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-D-phenylalanyl-2,4-dichloro-N-[(2S)-1-(4-fluorophenyl)-4-oxo-4-{[3-(pyridin-3-yl)propyl]amino}butan-2-yl]-D-phenylalaninamide, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Zack, A, Iketani, S, Williams, A, Vaz, D.R, Habashi, D.L, Choi, K, Resnick, S.J, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2023-09-30 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Development of small molecule non-covalent coronavirus 3CL protease inhibitors from DNA-encoded chemical library screening.
Nat Commun, 16, 2025
|
|
5N1K
 
 | |
6EYN
 
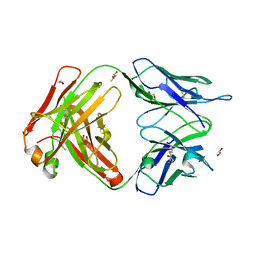 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
9HTZ
 
 | | M2-32, a new class A acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Ramos, J.L, Martinez-Rodriguez, S, Recio, M.I, de la Torre, J. | | Deposit date: | 2024-12-20 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermotolerant class A acid phosphatase active across broad pH range and diverse substrates.
Protein Sci., 34, 2025
|
|
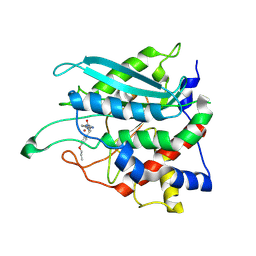
8XGB
 
 | |
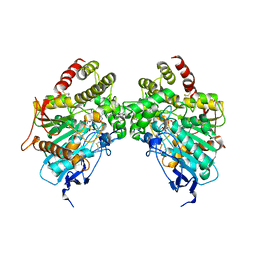
6O5S
 
 | |
8UWV
 
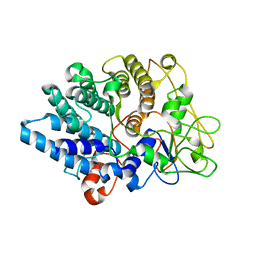 | |
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | Descriptor: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | Descriptor: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
9AV0
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
8V7U
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
8I6K
 
 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
8XF8
 
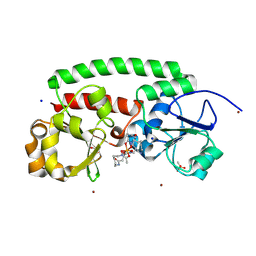 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine B | | Descriptor: | 1,2-ETHANEDIOL, Ferrioxamine B, Iron-hydroxamate ABC transporter substrate-binding protein FtsB, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 32, 2024
|
|
6O5R
 
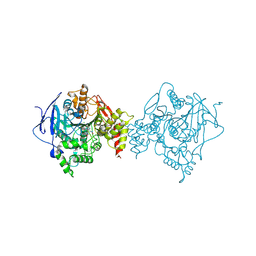 | |
9AV3
 
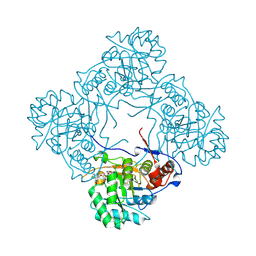 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUV
 
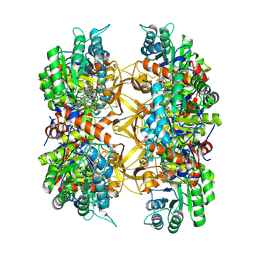 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
6XKZ
 
 | | R. capsulatus CIII2CIV tripartite super-complex, conformation B (SC-1B) | | Descriptor: | COPPER (II) ION, Cbb3-type cytochrome c oxidase subunit CcoP,Cytochrome c-type cyt cy, Cytochrome b, ... | | Authors: | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | Deposit date: | 2020-06-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
6OED
 
 | | CRYSTAL STRUCTURE OF THE RV144 C1-C2 SPECIFIC ANTIBODY CH55 FAB | | Descriptor: | CH55 Fab heavy chain, CH55 Fab light chain | | Authors: | Yan, F, Van, V, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Recognition Patterns of the C1/C2 Epitopes Involved in Fc-Mediated Response in HIV-1 Natural Infection and the RV114 Vaccine Trial.
Mbio, 11, 2020
|
|
6XT1
 
 | |
5LV5
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor LH1458 | | Descriptor: | 2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethyl-[[4-azanyl-1-(methoxymethyl)-2-oxidanylidene-pyrimidin-5-yl]methyl]-[(3~{S})-3-azanyl-4-oxidanyl-4-oxidanylidene-butyl]azanium, Protein arginine N-methyltransferase 6 | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
