8EKW
 
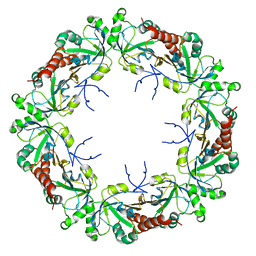 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8SS3
 
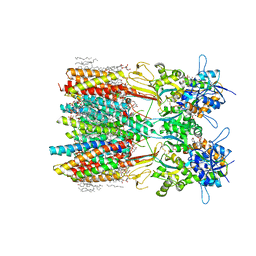 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS2
 
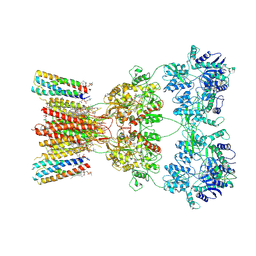 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6EAA
 
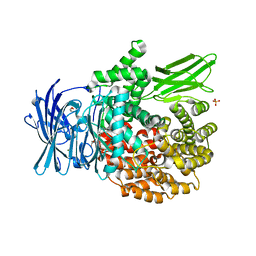 | |
8SS9
 
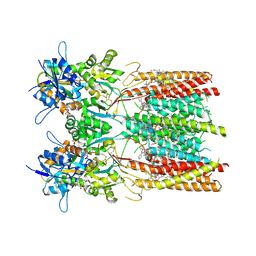 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4KWG
 
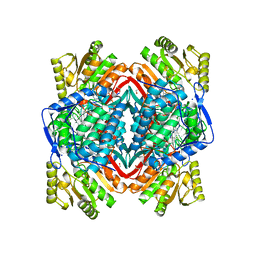 | | Crystal Structure Analysis of ALDH2+ALDiB13 | | Descriptor: | 1,2-ETHANEDIOL, 7-bromo-5-methyl-1H-indole-2,3-dione, Aldehyde dehydrogenase, ... | | Authors: | Hurley, T.D, Kimble-Hill, A.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones.
J.Med.Chem., 57, 2014
|
|
8SS6
 
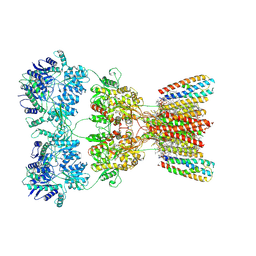 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4KZF
 
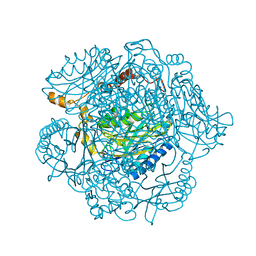 | | The mechanism of the amidases: The effect of the mutation E142L in the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase, CHLORIDE ION | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
8SDA
 
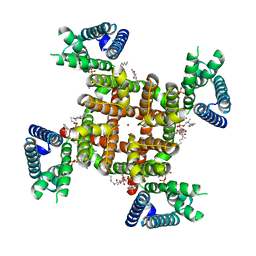 | | CryoEM structure of rat Kv2.1(1-598) L403A mutant in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
4KZK
 
 | |
8SD3
 
 | | CryoEM structure of rat Kv2.1(1-598) wild type in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
6EFC
 
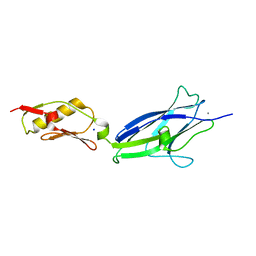 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EGU
 
 | |
6EH0
 
 | |
8SE6
 
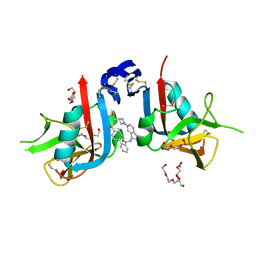 | | NKG2D complexed with inhibitor 36 | | Descriptor: | N-[(1S)-2-(dimethylamino)-2-oxo-1-{3-[3-(2,2,2-trifluoroethyl)azetidin-1-yl]phenyl}ethyl]-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein, TETRAETHYLENE GLYCOL, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Development of small molecule inhibitors of natural killer group 2D receptor (NKG2D).
Bioorg.Med.Chem.Lett., 96, 2023
|
|
6EHR
 
 | |
8EFY
 
 | |
4L4D
 
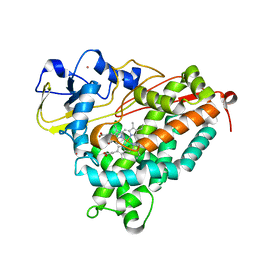 | | Structure of cyanide and camphor bound P450cam mutant L358A | | Descriptor: | CAMPHOR, CYANIDE ION, Camphor 5-monooxygenase, ... | | Authors: | Batabyal, D, Li, H, Poulos, T.L. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1.
Biochemistry, 52, 2013
|
|
6EJS
 
 | |
8EMT
 
 | | Cryo-EM analysis of the human aldehyde oxidase from liver | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Su, C, Lyu, M, Zhang, Z, Yu, E.W. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
4L58
 
 | |
6EKN
 
 | | Crystal structure of MMP12 in complex with inhibitor BE7. | | Descriptor: | (2~{S})-2-[2-[4-(4-methoxyphenyl)phenyl]sulfonylphenyl]pentanedioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ciccone, L, Tepshi, L, Nuti, E, Rossello, A, Stura, E.A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Development of Thioaryl-Based Matrix Metalloproteinase-12 Inhibitors with Alternative Zinc-Binding Groups: Synthesis, Potentiometric, NMR, and Crystallographic Studies.
J. Med. Chem., 61, 2018
|
|
6EKV
 
 | |
6EN0
 
 | |
8SRP
 
 | |
