5YAO
 
 | | The complex structure of SZ529 and expoxid | | Descriptor: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, SODIUM ION | | Authors: | Lian, W, Sun, Z.T, Zhou, J.H, Reetz, M.T. | | Deposit date: | 2017-09-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations
J. Am. Chem. Soc., 140, 2018
|
|
4I33
 
 | | Crystal structure of HCV NS3/4A R155K protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
4I26
 
 | | 2.20 Angstroms X-ray crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | 1,2-ETHANEDIOL, 2-aminomuconate 6-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Davis, I, Huo, L, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
3OE3
 
 | | Crystal structure of PliC-St, periplasmic lysozyme inhibitor of C-type lysozyme from Salmonella typhimurium | | Descriptor: | Putative periplasmic protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-12 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
4I31
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with compound 4 | | Descriptor: | (2R,6S,7E,10E,13aR,14aR,16aS)-2-{[7-methoxy-8-methyl-2-(propan-2-yloxy)quinolin-4-yl]oxy}-N-[(1-methylcyclopropyl)sulfonyl]-6-{[(1-methyl-1H-pyrazol-3-yl)carbonyl]amino}-5,16-dioxo-1,2,3,6,9,12,13,13a,14,15,16,16a-dodecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecine-14a(5H)-carboxamide, Genome polyprotein, HCV non-structural protein 4A, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-01-02 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9301 Å) | | Cite: | Molecular Mechanism by Which a Potent Hepatitis C Virus NS3-NS4A Protease Inhibitor Overcomes Emergence of Resistance.
J.Biol.Chem., 288, 2013
|
|
6CRB
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CF2, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(S)-{[(S)-[difluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR7
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHF, beta, gamma dATP analogue | | Descriptor: | 9-{2-deoxy-5-O-[(R)-{[(R)-[(R)-fluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-erythro-pentofuranosyl}-9H-purin-6-amine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR4
 
 | |
6CTL
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL-R/S isomers, beta, gamma dTTP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
5Y4A
 
 | |
2HIG
 
 | | Crystal Structure of Phosphofructokinase apoenzyme from Trypanosoma brucei. | | Descriptor: | 6-phospho-1-fructokinase, SODIUM ION | | Authors: | Martinez-Oyanedel, J, McNae, I.W, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Crystal Structure of Phosphofructokinase from a Eukaryote: Trypanosoma brucei.
J.Mol.Biol., 366, 2007
|
|
6CR8
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CHCL (R & S isomers), beta, gamma dATP analogue | | Descriptor: | 5'-O-[(R)-{[(R)-[(R)-chloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2'-deoxyadenosine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
2HLP
 
 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1INW
 
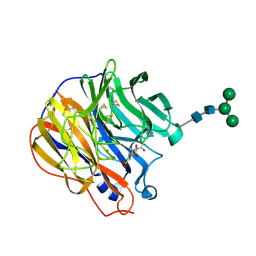 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1S)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
6CRC
 
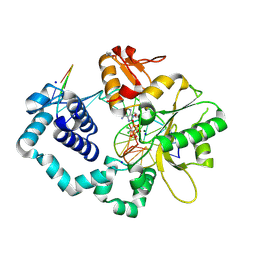 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CCL2, beta, gamma dATP analogue | | Descriptor: | 2'-deoxy-5'-O-[(R)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine, DNA polymerase beta, Downstream Primer Strand, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CTP
 
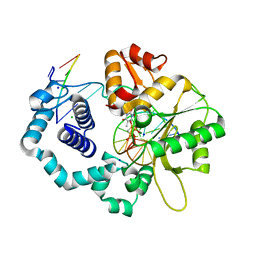 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CH2, beta, gamma dTTP analogue | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3'), ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CR9
 
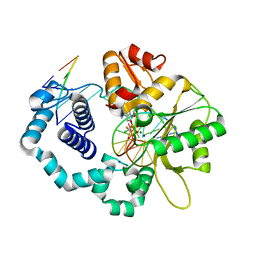 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CFCL, beta, gamma dATP analogue | | Descriptor: | 9-{5-O-[(R)-{[(R)-[(S)-chloro(fluoro)phosphonomethyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-deoxy-alpha-D-threo-pentofuranosyl}-9H-purin-6-amine, CHLORIDE ION, DNA polymerase beta, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
5YBZ
 
 | |
1INX
 
 | | A SIALIC ACID DERIVED PHOSPHONATE ANALOG INHIBITS DIFFERENT STRAINS OF INFLUENZA VIRUS NEURAMINIDASE WITH DIFFERENT EFFICIENCIES | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | White, C.L, Janakiraman, M.N, Laver, W.G, Philippon, C, Vasella, A, Air, G.M, Luo, M. | | Deposit date: | 1994-09-26 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A sialic acid-derived phosphonate analog inhibits different strains of influenza virus neuraminidase with different efficiencies.
J.Mol.Biol., 245, 1995
|
|
6CTV
 
 | | Ternary complex crystal structure of DNA polymerase Beta with a dideoxy terminated primer with CF2, beta, gamma dCTP analogue | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-deoxy-5'-O-[(R)-{[(R)-[difluoro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CHLORIDE ION, ... | | Authors: | Batra, V.K, Wilson, S.H. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping Functional Substrate-Enzyme Interactions in the pol beta Active Site through Chemical Biology: Structural Responses to Acidity Modification of Incoming dNTPs.
Biochemistry, 57, 2018
|
|
6CLZ
 
 | |
2R5O
 
 | | Crystal structure of the C-terminal domain of wzt | | Descriptor: | CHLORIDE ION, Putative ATP binding component of ABC-transporter, SODIUM ION, ... | | Authors: | Kimber, M.S, Cuthbertson, L, Whitfield, C. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate binding by a bacterial ABC transporter involved in polysaccharide export.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3PAK
 
 | | Crystal Structure of Rat Surfactant Protein A neck and carbohydrate recognition domain (NCRD) complexed with Mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, SODIUM ION, ... | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
6D8X
 
 | | PPAR gamma LBD complexed with the agonist GW1929 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CITRATE ANION, GLYCEROL, ... | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PPAR gamma LBD complexed with the agonist GW1929
To Be Published
|
|
5YUR
 
 | | DNA polymerase IV - DNA ternary complex 1 | | Descriptor: | DNA polymerase IV, DTN, SODIUM ION, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Nucleic Acids Res., 46, 2018
|
|
