8GL8
 
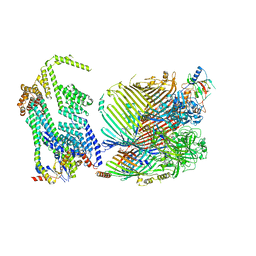 | |
8GLM
 
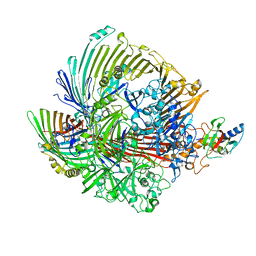 | |
8BJD
 
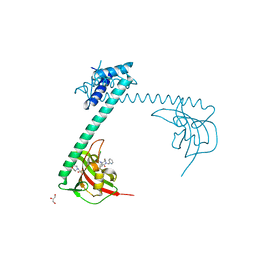 | | Full length structure of LpMIP with bound inhibitor JK095 | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BK5
 
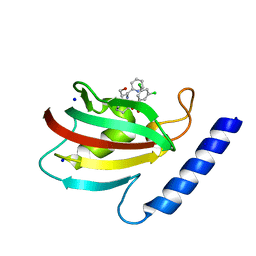 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJE
 
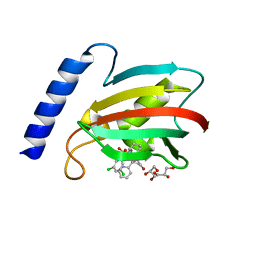 | | A structure of the truncated LpMIP with bound inhibitor JK236. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJC
 
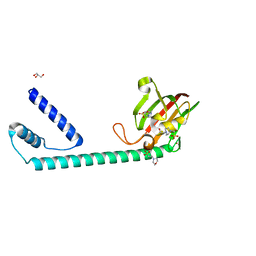 | | Full length structure of the apo-state LpMIP. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
7VML
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 1&2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMN
 
 | | Structure of recombinant RyR2 (EGTA dataset, class 2, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
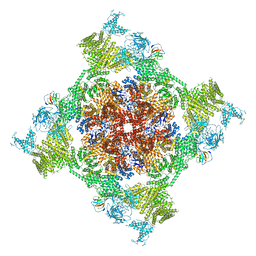 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
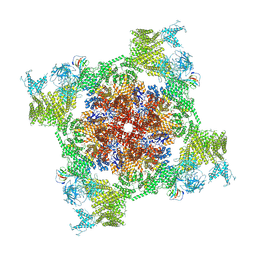 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMM
 
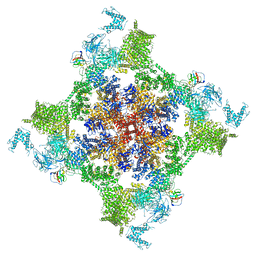 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMS
 
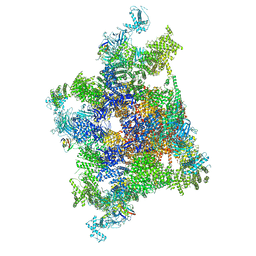 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
7VMR
 
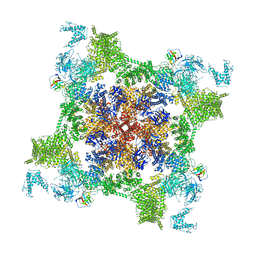 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
8JGA
 
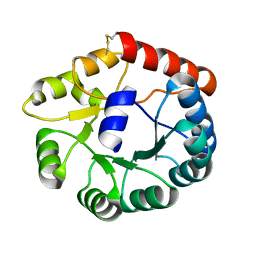 | | Cryo-EM structure of Mi3 fused with FKBP | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Zhang, H.W, Kang, W, Xue, C. | | Deposit date: | 2023-05-20 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Dynamic Metabolons Using Stimuli-Responsive Protein Cages.
J.Am.Chem.Soc., 146, 2024
|
|
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
8SEP
 
 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
2MPH
 
 | |
3PRD
 
 | |
