5WGG
 
 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, CteA, IRON/SULFUR CLUSTER, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-14 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
5WH8
 
 | | Cellulase Cel5C_n | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Koropatkin, N.M, Pope, P.B, Naas, A.E. | | Deposit date: | 2017-07-15 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | "Candidatus Paraporphyromonas polyenzymogenes" encodes multi-modular cellulases linked to the type IX secretion system.
Microbiome, 6, 2018
|
|
6SYQ
 
 | | ASR Alternansucrase in complex with isomaltotriose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-09-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
5WHZ
 
 | | PGDM1400-10E8v4 CODV Fab | | Descriptor: | Anti-HIV CODV-Fab Heavy chain, Anti-HIV CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.549 Å) | | Cite: | Trispecific broadly neutralizing HIV antibodies mediate potent SHIV protection in macaques.
Science, 358, 2017
|
|
4N2K
 
 | | Crystal structure of Protein Arginine Deiminase 2 (Q350A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5WIA
 
 | | Crystal structure of the segment, GNNSYS, from the low complexity domain of TDP-43, residues 370-375 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4N36
 
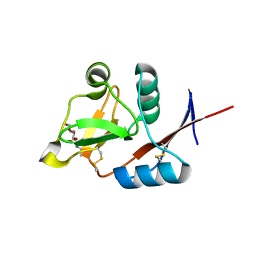 | | Structure of langerin CRD I313 D288 complexed with Me-GlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
4N3N
 
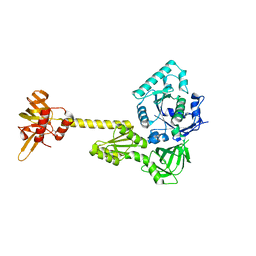 | |
4N43
 
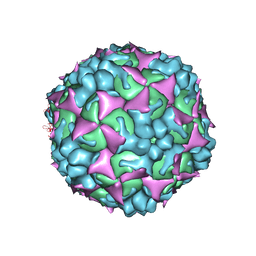 | |
5WKD
 
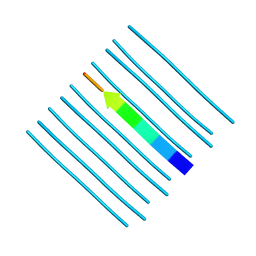 | | Crystal structure of the segment, GNNQGSN, from the low complexity domain of TDP-43, residues 300-306 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Guenther, E.L, Trinh, H, Sawaya, M.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WKF
 
 | | D30 TCR in complex with HLA-A*11:01-GTS1 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WKL
 
 | | 1.85 A resolution structure of MERS 3CL protease in complex with piperidine-based peptidomimetic inhibitor 17 | | Descriptor: | (1R,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[4-benzyl-1-(tert-butoxycarbonyl)piperidin-4-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element.
Eur J Med Chem, 150, 2018
|
|
4N0Z
 
 | |
4N6E
 
 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
5WKH
 
 | | D30 TCR in complex with HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS3 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
4N1T
 
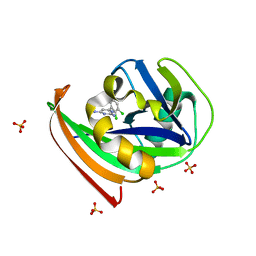 | | Structure of human MTH1 in complex with TH287 | | Descriptor: | 6-(2,3-dichlorophenyl)-N~4~-methylpyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
4N28
 
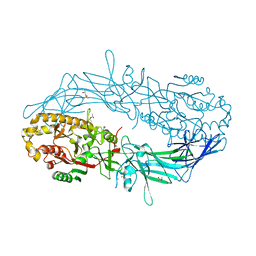 | | Crystal structure of Protein Arginine Deiminase 2 (1 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5WLH
 
 | |
5WLU
 
 | |
5WM4
 
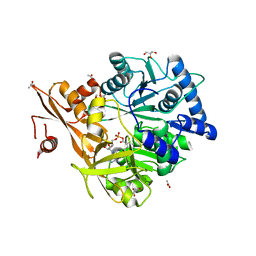 | | Crystal Structure of CahJ in Complex with 6-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-6-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WMO
 
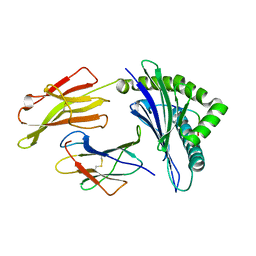 | | Crystal Structure of HLA-B7 in complex with RPP, an EBV peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses.
J. Immunol., 200, 2018
|
|
5WN0
 
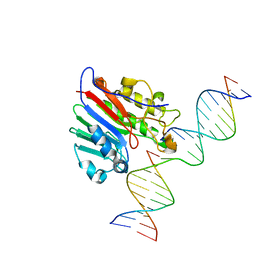 | | APE1 exonuclease substrate complex with a C/G match | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
5WNS
 
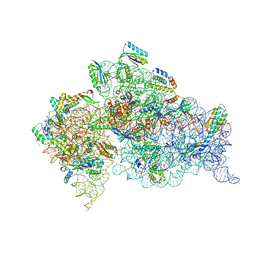 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WN9
 
 | | Structure of antibody 2D10 bound to the central conserved region of RSV G | | Descriptor: | Major surface glycoprotein G, scFv 2D10 | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
5WNN
 
 | |
