5W1I
 
 | |
6T16
 
 | | ASR Alternansucrase in complex with panose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Cioci, G, Molina, M, Moulis, C, Remaud-Simeon, M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation.
J.Biol.Chem., 295, 2020
|
|
5VZP
 
 | | Sperm whale myoglobin H64Q with nitric oxide | | Descriptor: | GLYCEROL, Myoglobin, NITRIC OXIDE, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
5VZX
 
 | | Crystal structure of crenezumab Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Crenezumab Fab heavy chain, ... | | Authors: | Ultsch, M, Wang, W. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Sci Rep, 6, 2016
|
|
4N1C
 
 | | Structural evidence for antigen receptor evolution | | Descriptor: | Lysozyme C, immunoglobulin variable light chain domain | | Authors: | Langley, D.B, Rouet, R, Roome, B, Stock, D, Christ, D. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural reconstruction of protein ancestry.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W21
 
 | | Crystal Structure of a 1:1:1 FGF23-FGFR1c-aKlotho Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibroblast growth factor 23, Fibroblast growth factor receptor 1, ... | | Authors: | Mohammadi, M. | | Deposit date: | 2017-06-05 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling.
Nature, 553, 2018
|
|
5W2S
 
 | |
4N24
 
 | | Crystal structure of Protein Arginine Deiminase 2 (100 uM Ca2+) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4MRU
 
 | |
4N2M
 
 | | Crystal structure of Protein Arginine Deiminase 2 (E354A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
5W6C
 
 | | UCA Fab (unbound) from 6649 Lineage | | Descriptor: | UCA Fab heavy chain, UCA Fab light chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2017-06-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VXF
 
 | |
4N3A
 
 | |
4N41
 
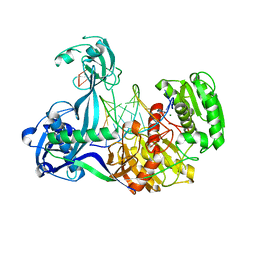 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | Descriptor: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5W0N
 
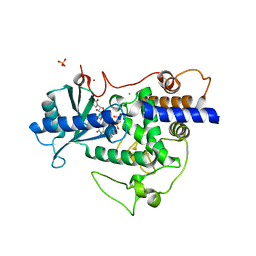 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
5W4L
 
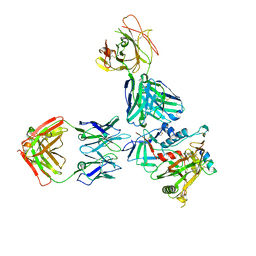 | | Crystal structure of the non-neutralizing and ADCC-potent C11-like antibody N12-i3 in complex with HIV-1 clade A/E gp120, the CD4 mimetic M48U1, and the antibody N5-i5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody N12-i3 Fab heavy chain, Antibody N12-i3 light chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Targeting the Late Stage of HIV-1 Entry for Antibody-Dependent Cellular Cytotoxicity: Structural Basis for Env Epitopes in the C11 Region.
Structure, 25, 2017
|
|
5W5W
 
 | |
4N6F
 
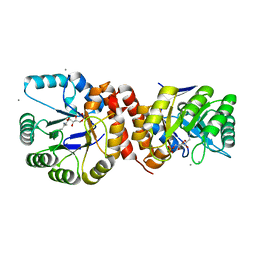 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | Descriptor: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4N71
 
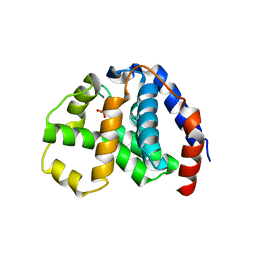 | | X-Ray Crystal Structure of 2-amino-1-hydroxyethylphosphonate-bound PhnZ | | Descriptor: | FE (III) ION, Predicted HD phosphohydrolase PhnZ, [(1R)-2-amino-1-hydroxyethyl]phosphonic acid | | Authors: | Woersdoerfer, B, Lingaraju, M, Yennawar, N, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.-E. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5W76
 
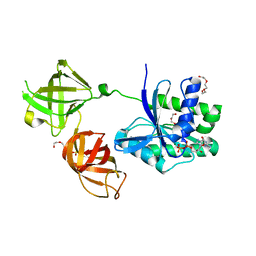 | | Crystal Structure of Reconstructed Bacterial Elongation Factor Node 168 | | Descriptor: | Ancestral Elogation Factor N153, DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2017-06-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural and Dynamics Comparison of Thermostability in Ancient, Modern, and Consensus Elongation Factor Tus.
Structure, 26, 2018
|
|
5W7F
 
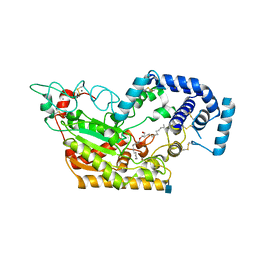 | | Murine acyloxyacyl hydrolase (AOAH), S262A mutant, with lipid A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the mammalian lipopolysaccharide detoxifier.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4MZZ
 
 | |
5W37
 
 | |
5W3R
 
 | | SH2B1 SH2 Domain | | Descriptor: | PHENOL, PHOSPHATE ION, SH2B adapter protein 1 | | Authors: | McKercher, M.A, Wuttke, D.S. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Diversity in peptide recognition by the SH2 domain of SH2B1.
Proteins, 86, 2018
|
|
