2X8T
 
 | | Crystal Structure of the Abn2 H318A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | deSanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
5QJI
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1899842917 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]-5-methyl-1,3,4-thiadiazol-2(3H)-one, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2L7J
 
 | |
2LM5
 
 | |
4GY9
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5QDY
 
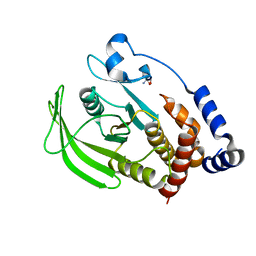 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000599c | | Descriptor: | 1-methyl-3-(thiophen-2-yl)-1H-pyrazol-5-amine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
2WM0
 
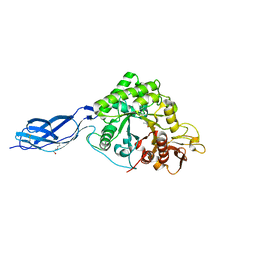 | | Chitinase A from Serratia marcescens ATCC990 in complex with Chitobio- thiazoline thioamide. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Taylor, E.J, Dennis, R.J, Macdonald, J.M, Tarling, C.A, Knapp, S, Withers, S.G, Davies, G.J. | | Deposit date: | 2009-06-29 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chitinase Inhibition by Chitobiose and Chitotriose Thiazolines.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3WKY
 
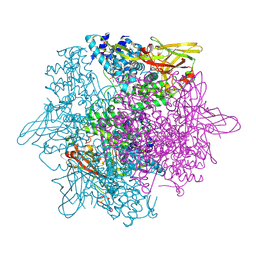 | | Crystal structure of hemolymph type prophenoloxidase (proPOb) from crustacean | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Masuda, T, Mikami, B. | | Deposit date: | 2013-11-02 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The crystal structure of a crustacean prophenoloxidase provides a clue to understanding the functionality of the type 3 copper proteins.
Febs J., 281, 2014
|
|
5QK1
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z2027049478 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1S2P
 
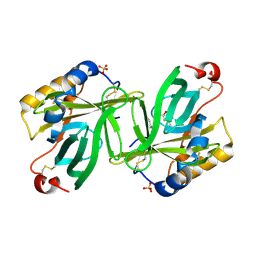 | | The structure and refinement of apocrustacyanin C2 to 1.3A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Crustacyanin C2 subunit, SULFATE ION | | Authors: | Habash, J, Helliwell, J.R, Raftery, J, Cianci, M, Rizkallah, P.J, Chayen, N.E, NNeji, G.A, Zakalsky, P.F. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure and refinement of apocrustacyanin C2 to 1.3 A resolution and the search for differences between this protein and the homologous apoproteins A1 and C1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2WXV
 
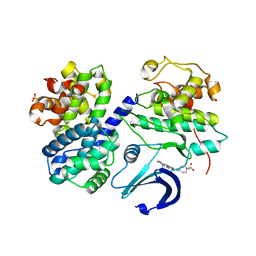 | | Structure of CDK2-CYCLIN A with a Pyrazolo(4,3-h) quinazoline-3- carboxamide inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1-DIMETHYL-8-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Traquandi, G, Ciomei, M, Ballinari, D, Casale, E, Colombo, N, Croci, V, Fiorentini, F, Isacchi, A, Longo, A, Mercurio, C, Panzeri, A, Pastori, W, Pevarello, P, Volpi, D, Roussel, P, Vulpetti, A, Brasca, M.G. | | Deposit date: | 2009-11-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Potent Pyrazolo[4,3-H]Quinazoline-3-Carboxamides as Multi-Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
7E9L
 
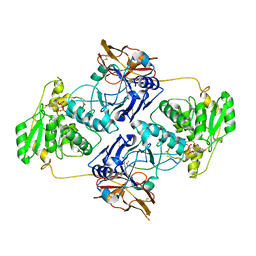 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man short peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
7E9K
 
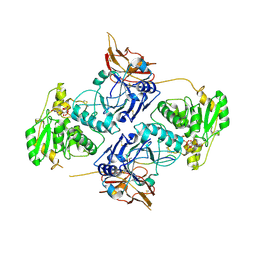 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man long peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
1WU6
 
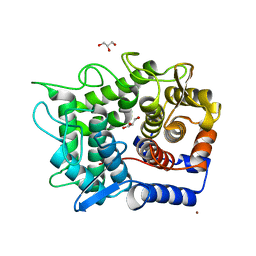 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase E70A mutant complexed with xylobiose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1GOO
 
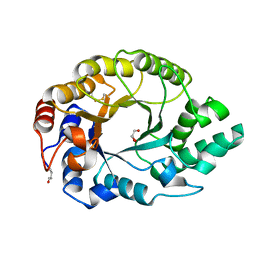 | | Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex | | Descriptor: | ENDO-1,4-BETA-XYLANASE, GLYCEROL | | Authors: | Eckert, K, Andrei, C, Larsen, S, Lo Leggio, L. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Substrate Specificity and Subsite Mobility in T. Aurantiacus Xylanase 10A
FEBS Lett., 509, 2001
|
|
4HAY
 
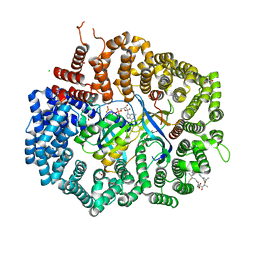 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K548E,K579Q)-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3P0A
 
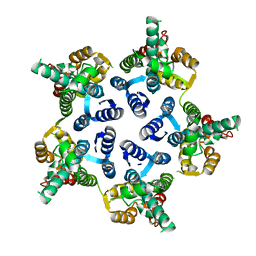 | |
4HC6
 
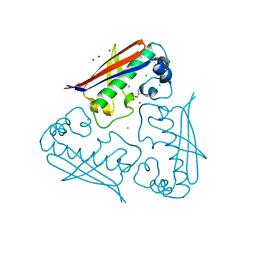 | | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, H.-B, Han, G.-W, Hung, L.-W, Terwilliger, C.T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution
To be Published
|
|
3HJ1
 
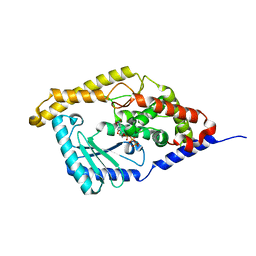 | | Minor Editosome-Associated TUTase 1 with bound UTP | | Descriptor: | Minor Editosome-Associated TUTase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Stagno, J, Luecke, H. | | Deposit date: | 2009-05-20 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Mitochondrial Editosome-Like Complex Associated TUTase 1 Reveals Divergent Mechanisms of UTP Selection and Domain Organization.
J.Mol.Biol., 399, 2010
|
|
3OIK
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3,17-O,O-bis-sulfamate | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-ETHYLESTRA-1(10),2,4-TRIENE-3,17-DIYL DISULFAMATE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
4DK8
 
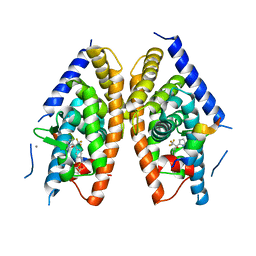 | | Crystal structure of LXR ligand binding domain in complex with partial agonist 5 | | Descriptor: | ACETATE ION, CALCIUM ION, N-methyl-N-(4-{(1S)-2,2,2-trifluoro-1-hydroxy-1-[1-(2-methoxyethyl)-1H-pyrrol-2-yl]ethyl}phenyl)benzenesulfonamide, ... | | Authors: | Piper, D.E, Kopecky, D.J, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3HL3
 
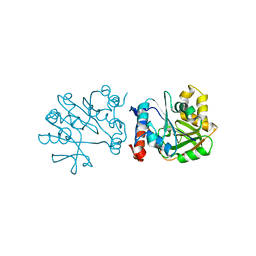 | | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose. | | Descriptor: | Glucose-1-phosphate thymidylyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | 2.76 Angstrom Crystal Structure of a Putative Glucose-1-Phosphate Thymidylyltransferase from Bacillus anthracis in Complex with a Sucrose.
TO BE PUBLISHED
|
|
1H95
 
 | | Solution structure of the single-stranded DNA-binding Cold Shock Domain (CSD) of human Y-box protein 1 (YB1) determined by NMR (10 lowest energy structures) | | Descriptor: | Y-BOX BINDING PROTEIN | | Authors: | Kloks, C.P.A.M, Spronk, C.A.E.M, Hoffmann, A, Vuister, G.W, Grzesiek, S, Hilbers, C.W. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
J.Mol.Biol., 316, 2002
|
|
5QK8
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1271660837 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1UCF
 
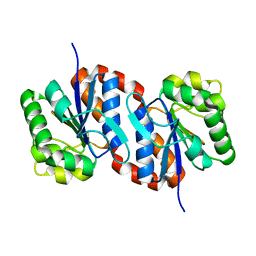 | | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Honbou, K, Suzuki, N.N, Horiuchi, M, Niki, T, Taira, T, Ariga, H, Inagaki, F. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of DJ-1, a Protein Related to Male Fertility and Parkinson's Disease
J.BIOL.CHEM., 278, 2003
|
|
