5I3M
 
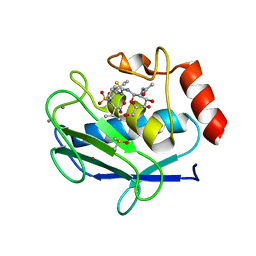 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated thiourea-linked carboxylate zinc-chelator water-soluble inhibitor (DC31). | | Descriptor: | (2S)-2-{[2-({[(2R,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]carbamothioyl}amino)ethyl](biphenyl-4-ylsulfonyl)amino}-3-methylbutanoic acid (non-preferred name), 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
6CV0
 
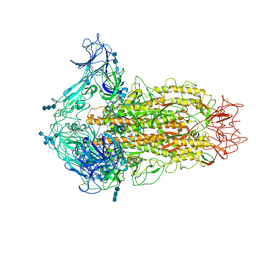 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
6SU4
 
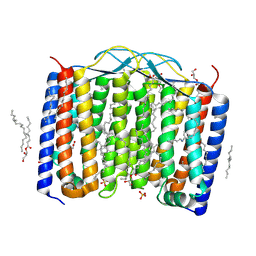 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6CED
 
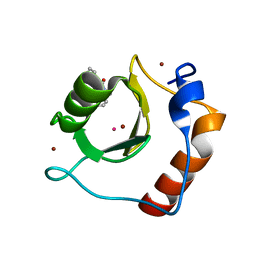 | | Crystal structure of fragment 3-(3-Methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methyl-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
5O8X
 
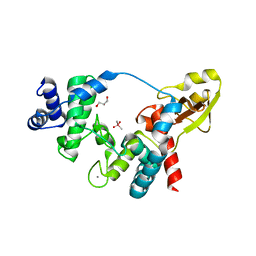 | |
6CFL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with lysyl-CAM and bound to protein Y (YfiA) at 2.6A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
6J2Q
 
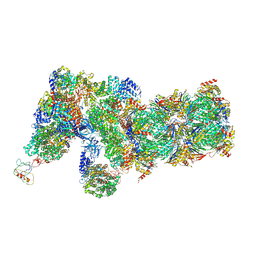 | | Yeast proteasome in Ub-accepted state (C1-b) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Cong, Y. | | Deposit date: | 2019-01-02 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
4RVO
 
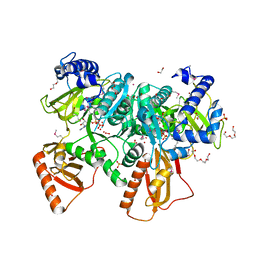 | |
6J5J
 
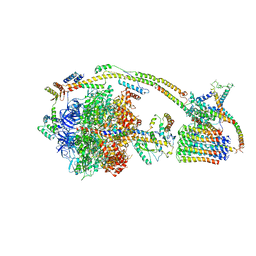 | | Cryo-EM structure of the mammalian E-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6CE6
 
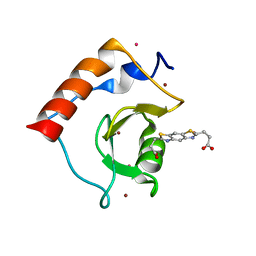 | | Structure of HDAC6 zinc-finger ubiquitin binding domain soaked with 3,3'-(benzo[1,2-d:5,4-d']bis(thiazole)-2,6-diyl)dipropionic acid | | Descriptor: | 3,3'-(benzo[1,2-d:5,4-d']bis[1,3]thiazole-2,6-diyl)dipropanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6SL5
 
 | | Dunaliella Photosystem I Supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Caspy, I, Malavath, T, Klaiman, D, Shkolinsky, Y. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and energy transfer pathways of the Dunaliella Salina photosystem I supercomplex.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
5O60
 
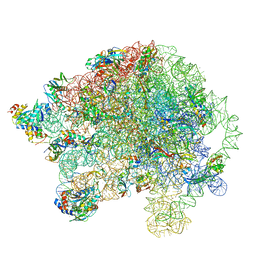 | | Structure of the 50S large ribosomal subunit from Mycobacterium smegmatis | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
5EAI
 
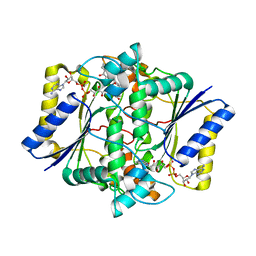 | | Crystal Structure of NAD(P)H dehydrogenase, quinone 1 complexed with a chemotherapeutic naphthoquinone E6a | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{S})-3-bromanyl-1,4-bis(oxidanylidene)-2,3-dihydronaphthalen-2-yl]-3-oxidanyl-2,3-dihydronaphthalene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Pidugu, L.S, Mbimba, J.E, Ahmad, M, Pozharski, E, Sausville, E.A, Emadi, A, Toth, E.A. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A direct interaction between NQO1 and a chemotherapeutic dimeric naphthoquinone.
Bmc Struct.Biol., 16, 2016
|
|
6JCV
 
 | | Cryo-EM structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH7.5 | | Descriptor: | MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
4ZHH
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RZ8
 
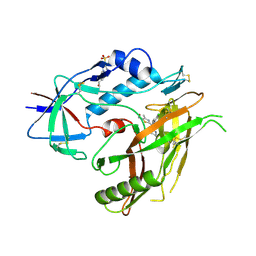 | | Crystal structure of HIV-1 gp120 core in complex with NBD-11021, a small molecule CD4-antagonist | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-chlorophenyl)-N-{(S)-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl][(2R)-piperidin-2-yl]methyl}-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of a Small Molecule CD4-Antagonist with Broad Spectrum Anti-HIV-1 Activity.
J.Med.Chem., 58, 2015
|
|
6SRO
 
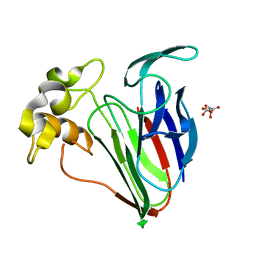 | |
5FDU
 
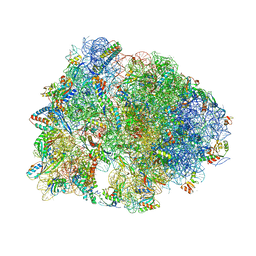 | | Crystal structure of the Metalnikowin I antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
4ZGD
 
 | | Mutant R157A of Fe-Type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit | | Authors: | Wu, R, Martinez, S, Holz, R, Liu, D. | | Deposit date: | 2015-04-22 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analyzing the catalytic role of active site residues in the Fe-type nitrile hydratase from Comamonas testosteroni Ni1.
J.Biol.Inorg.Chem., 20, 2015
|
|
5OHS
 
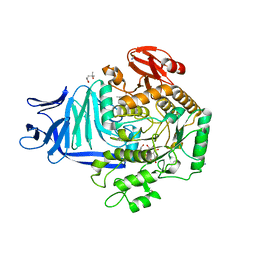 | | A GH31 family sulfoquinovosidase mutant D455N in complex with pNPSQ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-nitrophenyl alpha-D-6-sulfoquinovoside, ... | | Authors: | Jin, Y, Williams, S.J, Goddard-Borger, E, Davies, G.J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biochemical Insights into the Function and Evolution of Sulfoquinovosidases.
ACS Cent Sci, 4, 2018
|
|
6CE8
 
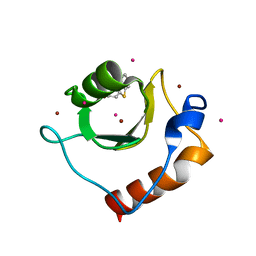 | | Crystal structure of fragment 2-(Benzo[d]thiazol-2-yl)acetic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | (1,3-benzothiazol-2-yl)acetic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEC
 
 | | Crystal structure of fragment 3-(3-Methoxy-2-quinoxalinyl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEF
 
 | | Crystal structure of fragment 3-(1,3-Benzothiazol-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
4RUK
 
 | |
6H4N
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
