3Q1Y
 
 | |
3QQP
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1VBE
 
 | | POLIOVIRUS (TYPE 3, SABIN STRAIN, MUTANT 242-H2) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
4CS4
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, 2-{[dihydroxy(4-aminoethylphenyl)-{4}-sulfanyl]amino}-3-hydroxypropanoic acid, MAGNESIUM ION, ... | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4CS2
 
 | | Catalytic domain of Pyrrolysyl-tRNA synthetase mutant Y306A, Y384F in its apo form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PYRROLYSINE--TRNA LIGASE | | Authors: | Schmidt, M.J, Weber, A, Pott, M, Welte, W, Summerer, D. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Furan-Amino Acid Recognition by a Polyspecific Aminoacyl-tRNA-Synthetase and its Genetic Encoding in Human Cells.
Chembiochem, 15, 2014
|
|
4BJY
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Platinum derivative | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BJZ
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Native data | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BAB
 
 | | Redesign of a Phenylalanine Aminomutase into a beta-Phenylalanine Ammonia Lyase | | Descriptor: | PHENYLALANINE AMINOMUTASE | | Authors: | Bartsch, S, Wybenga, G.G, Jansen, M, Heberling, M.M, Wu, B, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2012-09-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Redesign of a Phenylalanine Aminomutase Into a Phenylalanine Ammonia Lyase
To be Published
|
|
4BK1
 
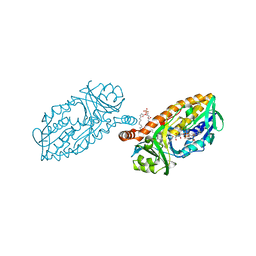 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: H213S mutant in complex with 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
1XE4
 
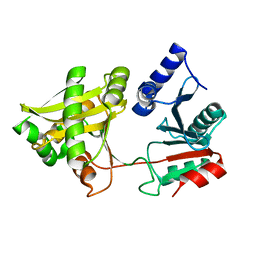 | | Crystal Structure of Weissella viridescens FemX (K36M) Mutant | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Arthur, M, Mayer, C. | | Deposit date: | 2004-09-09 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of the UDP-MurNAc-Pentapeptide-Binding Cavity of the FemX Alanyl Transferase from Weissella viridescens
J.Bacteriol., 187, 2005
|
|
1XFH
 
 | |
1XC1
 
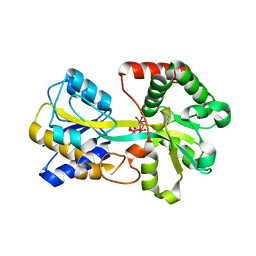 | | Oxo Zirconium(IV) Cluster in the Ferric Binding Protein (FBP) | | Descriptor: | OXO ZIRCONIUM(IV) CLUSTER, periplasmic iron-binding protein | | Authors: | Zhong, W, Alexeev, D, Harvey, I, Guo, M, Hunter, D.J.B, Zhu, H, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assembly of an Oxo-Zirconium(IV) Cluster in a Protein Cleft
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1VBA
 
 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R78206 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE PROPYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
4D7E
 
 | | An unprecedented NADPH domain conformation in Lysine Monooxygenase NbtG from Nocardia farcinica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-LYS MONOOXYGENASE | | Authors: | Binda, C, Robinson, R, Keul, N, Rodriguez, P, Robinson, H.H, Mattevi, A, Sobrado, P. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Unprecedented Nadph Domain Conformation in Lysine Monooxygenase Nbtg Provides Insights Into Uncoupling of Oxygen Consumption from Substrate Hydroxylation.
J.Biol.Chem., 290, 2015
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3RLK
 
 | |
1XIX
 
 | |
5TNH
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 17,18-EpETE hydrolysis intermediate | | Descriptor: | (5Z,8Z,11Z,14Z,17R,18R)-17,18-dihydroxyicosa-5,8,11,14-tetraenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNS
 
 | |
5TNF
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 19,20-EpDPE hydrolysis intermediate | | Descriptor: | (4Z,7Z,10Z,13Z,16Z,19R,20R)-19,20-dihydroxydocosa-4,7,10,13,16-pentaenoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
5TNQ
 
 | |
5TNN
 
 | |
3OPZ
 
 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | Authors: | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
5TNL
 
 | |
5TND
 
 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 1,2-Epoxycyclohexane hydrolysis intermediate | | Descriptor: | (1R,2R)-cyclohexane-1,2-diol, (1S,2S)-cyclohexane-1,2-diol, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-10-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.
Structure, 25, 2017
|
|
