8BP8
 
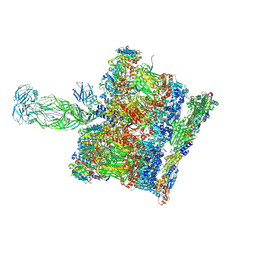 | | SPA of Trypsin untreated Rotavirus TLP spike | | Descriptor: | CALCIUM ION, Inner capsid protein VP2, Intermediate capsid protein VP6, ... | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
8B6F
 
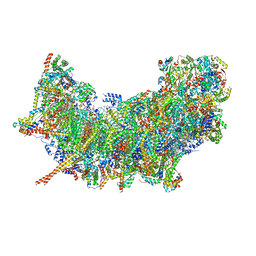 | | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
6ZKF
 
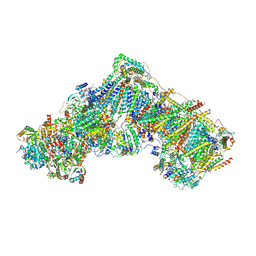 | | Complex I during turnover, open3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-14 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
8Y6O
 
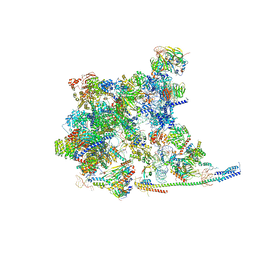 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
6ZKU
 
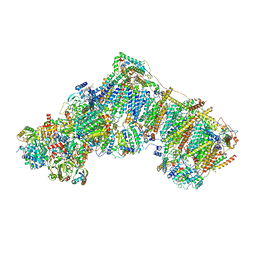 | | Deactive complex I, open3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
8B6H
 
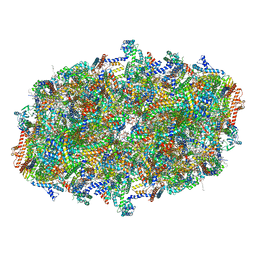 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
7EE6
 
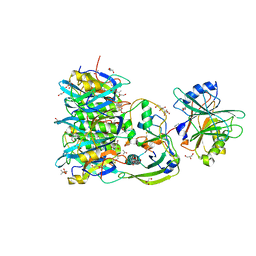 | | Crystal structure of PltC toxin | | Descriptor: | ACETONE, CITRATE ANION, Cytolethal distending toxin subunit B family protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
6UZV
 
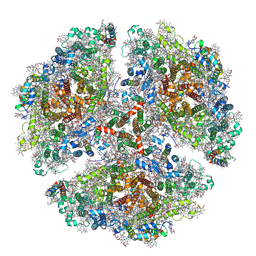 | | The structure of a red shifted photosystem I complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of a red-shifted photosystem I reveals a red site in the core antenna.
Nat Commun, 11, 2020
|
|
6VFD
 
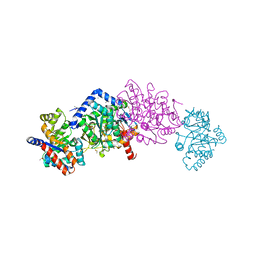 | | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site.
To be Published
|
|
8B6J
 
 | | Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apocytochrome b, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
6V22
 
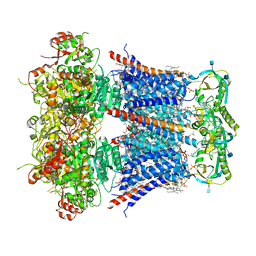 | | Cryo-EM structure of Ca2+-bound hsSlo1-beta4 channel complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
8XXL
 
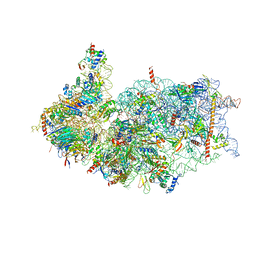 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
5LEX
 
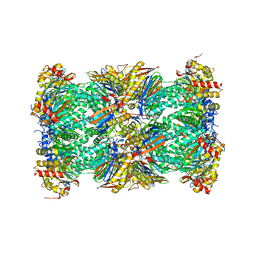 | | Native human 20S proteasome in Mg-Acetate at 2.2 Angstrom | | Descriptor: | MAGNESIUM ION, PENTAETHYLENE GLYCOL, POTASSIUM ION, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
7DUI
 
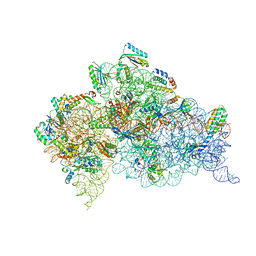 | |
8Y1E
 
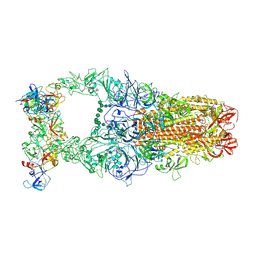 | | 3up-TM conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
7DUG
 
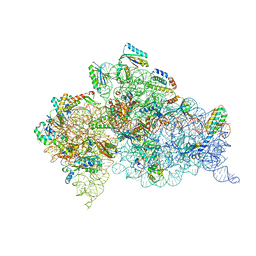 | |
8Y1C
 
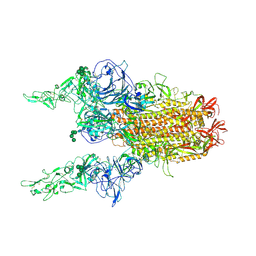 | | 2up-1 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
7DUA
 
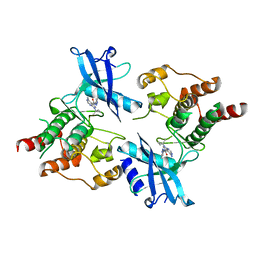 | |
8Y19
 
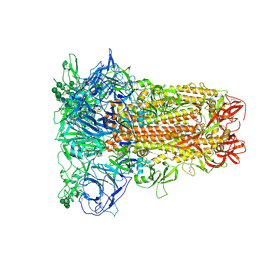 | | Closed conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1G
 
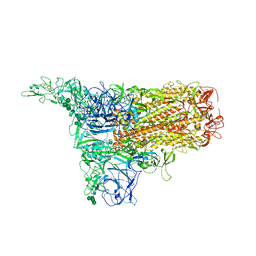 | | The 1up conformation of the HKU1-B S protein in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8Y1A
 
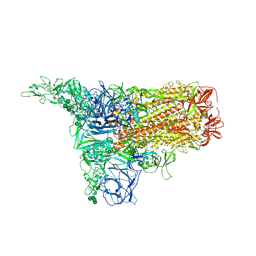 | | 1up-1 conformation of HKU1-B S protein after incubation of the receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the recognition of HCoV-HKU1 by human TMPRSS2.
Cell Res., 34, 2024
|
|
8BUU
 
 | | ARE-ABCF VmlR2 bound to a 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2022-11-30 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR.
Nucleic Acids Res., 51, 2023
|
|
8XLM
 
 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-12-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
7DUK
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''MS bound | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 30S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H, Destan, E. | | Deposit date: | 2021-01-09 | | Release date: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N1,3''MS bound
to be published
|
|
