7VS2
 
 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
7TZJ
 
 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
6HRO
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118a | | Descriptor: | 1-[2-[4-[4-(4-chlorophenyl)-3-methyl-1~{H}-pyrazol-5-yl]-3-oxidanyl-phenoxy]ethyl]piperidin-1-ium-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
7UEH
 
 | | Pyruvate kinase from Zymomonas mobilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pyruvate kinase | | Authors: | McFarlane, J.S, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.4 angstrom structure of Zymomonas mobilis pyruvate kinase: Implications for stability and regulation.
Arch.Biochem.Biophys., 744, 2023
|
|
6QZ1
 
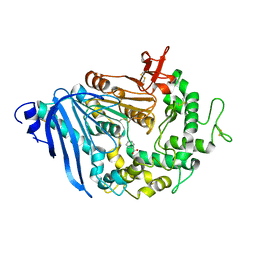 | | Structure of MHETase from Ideonella sakaiensis | | Descriptor: | BENZOIC ACID, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase | | Authors: | Allen, M.D, Johnson, C.W, Knott, B.C, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2019-03-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a two-enzyme system for plastics depolymerization.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7UMJ
 
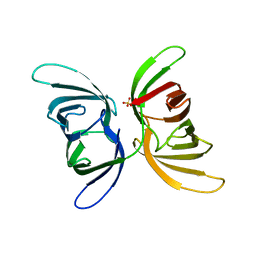 | | Crystal structure of recombinant Solieria filiformis lectin (rSfL) | | Descriptor: | Lectin SfL-1, SULFATE ION | | Authors: | Chaves, R.P, Bezerra, E.H.S, da Silva, F.M.S, Carneiro, R.F, Sampaio, A.H, Rocha, B.A.M, Nagano, C.S. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural study and antimicrobial and wound healing effects of lectin from Solieria filiformis (Kutzing) P.W.Gabrielson.
Biochimie, 214, 2023
|
|
6I3B
 
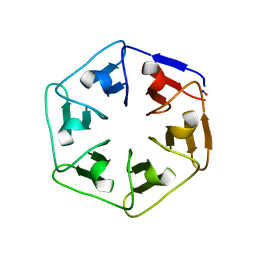 | |
7YXV
 
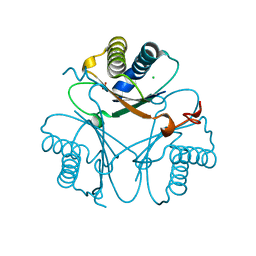 | |
4P2T
 
 | |
8ZC6
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.85 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
4P7A
 
 | | Crystal Structure of human MLH1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Mlh1, MAGNESIUM ION, ... | | Authors: | Tempel, W, Lam, R, Zeng, H, Walker, J.R, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBZ
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
6LD1
 
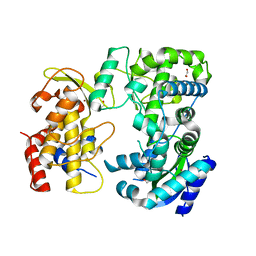 | | Zika NS5 polymerase domain | | Descriptor: | 1,2-ETHANEDIOL, RNA-directed RNA polymerase NS5, ZINC ION | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
8UMC
 
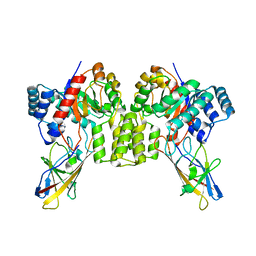 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), cryo-EM | | Descriptor: | DUF6379 domain-containing protein, MANGANESE (II) ION, Xylose isomerase-like TIM barrel domain-containing protein | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
6KZ8
 
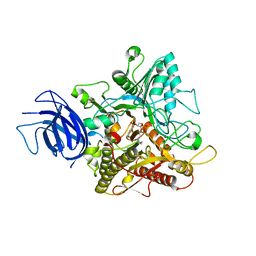 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
8ZBY
 
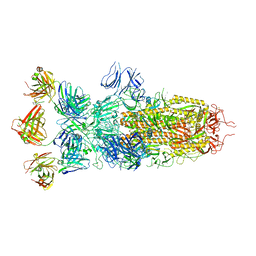 | | SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC1
 
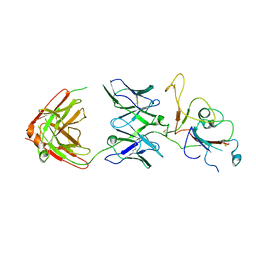 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC4
 
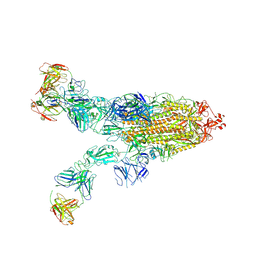 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
6L3X
 
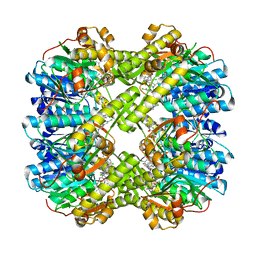 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1~{R})-1-[[(2~{S})-2-[[2,5-bis(chloranyl)phenyl]carbonylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-methyl-butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3054 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
6LD5
 
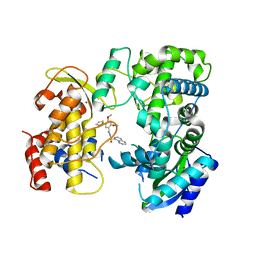 | | Zika NS5 polymerase domain | | Descriptor: | 2,4-dimethoxy-5-thiophen-2-yl-benzoic acid, RNA-directed RNA polymerase NS5, ZINC ION, ... | | Authors: | El Sahili, A, Lescar, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.
J.Virol., 94, 2020
|
|
6KZ9
 
 | | Crystal structure of plant Phospholipase D alpha | | Descriptor: | CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6LGT
 
 | | Complex structure of HPPD with an inhibitor Y16542 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chlorophenyl)-6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.Y, Yang, W.C, Yang, G.F. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Discovery of Novel Pyrazole-Quinazoline-2,4-dione Hybrids as 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 68, 2020
|
|
