7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL6
 
 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
5JKL
 
 | |
7BGN
 
 | |
8UHI
 
 | | Structure of the far-red light-absorbing allophycocyanin core expressed during FaRLiP | | Descriptor: | MAGNESIUM ION, RIBULOSE-1,5-DIPHOSPHATE, Ribulose bisphosphate carboxylase large subunit, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W, Shen, G. | | Deposit date: | 2023-10-09 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structure of the antenna complex expressed during far-red light photoacclimation in Synechococcus sp. PCC 7335.
J.Biol.Chem., 300, 2023
|
|
6WY0
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-40 A.K.A 7-[(1R)-1-phenyl-3-{[(1r,4r)-4-phenylcyclohexyl]amino}propyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-{(1R)-1-phenyl-3-[(trans-4-phenylcyclohexyl)amino]propyl}-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
6U0S
 
 | | Crystal structure of the flavin-dependent monooxygenase PieE in complex with FAD and substrate | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, 2-[(2E,5E,7E,9R,10R,11E)-10-hydroxy-3,7,9,11-tetramethyltrideca-2,5,7,11-tetraen-1-yl]-6-methoxy-3-methylpyridin-4-ol, CHLORIDE ION, ... | | Authors: | Shi, R, Manenda, M. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
7B93
 
 | | Cryo-EM structure of mitochondrial complex I from Mus musculus inhibited by IACS-2858 at 3.0 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1-[[3-(4-methylsulfonylpiperidin-1-yl)phenyl]methyl]-5-[3-[4-(trifluoromethyloxy)phenyl]-1,2,4-oxadiazol-5-yl]pyridin-2-one, ... | | Authors: | Chung, I, Hirst, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cork-in-bottle mechanism of inhibitor binding to mammalian complex I.
Sci Adv, 7, 2021
|
|
8UOX
 
 | | Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Johnson, S, Deme, J.C, Lea, S.M. | | Deposit date: | 2023-10-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
6U2J
 
 | | EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
8UFB
 
 | |
6WJN
 
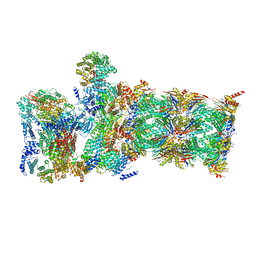 | |
6WML
 
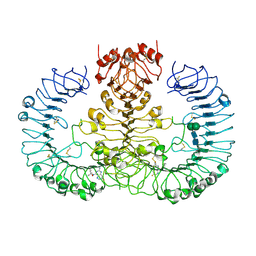 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
6WN6
 
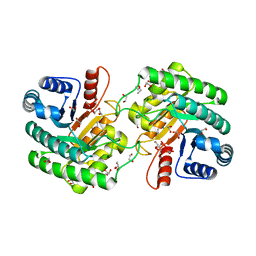 | | Crystal structure of 3-keto-D-glucoside 4-epimerase, YcjR, from E. coli, apo form | | Descriptor: | 1,2-ETHANEDIOL, 3-keto-D-glucoside 4-epimerase, MANGANESE (II) ION | | Authors: | Mabanglo, M.F, Raushel, F.M, Mukherjee, K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Reaction Mechanism of YcjR, an Epimerase That Facilitates the Interconversion of d-Gulosides to d-Glucosides inEscherichia coli.
Biochemistry, 59, 2020
|
|
7YQ2
 
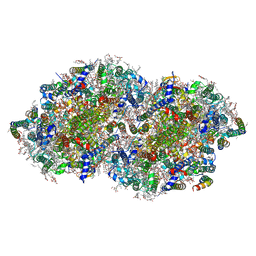 | | Crystal structure of photosystem II expressing psbA2 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
7YQ7
 
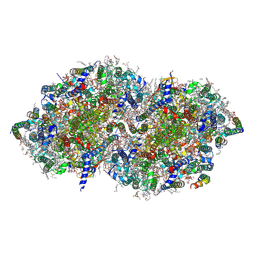 | | Crystal structure of photosystem II expressing psbA3 gene only | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nakajima, Y, Suga, M, Shen, J.R. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of photosystem II from a cyanobacterium expressing psbA 2 in comparison to psbA 3 reveal differences in the D1 subunit.
J.Biol.Chem., 298, 2022
|
|
5IY7
 
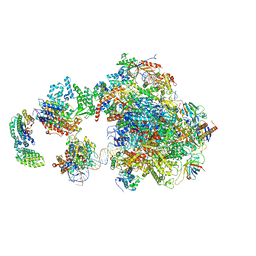 | | Human holo-PIC in the open state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
6TWY
 
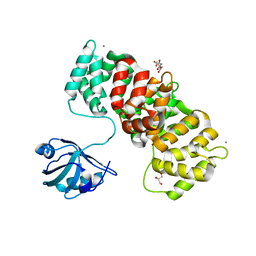 | | MAGI1_2 complexed with a phosphomimetic RSK1 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6WH3
 
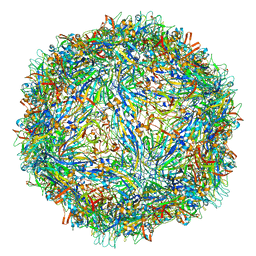 | | Capsid structure of Penaeus monodon metallodensovirus at pH 8.2 | | Descriptor: | CALCIUM ION, Penaeus monodon metallodensovirus major capsid protein | | Authors: | Penzes, J.J, Pham, H.T, Chipman, P, Bhattacharya, N, McKenna, R, Agbandje-McKenna, M, Tijssen, P. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BOI
 
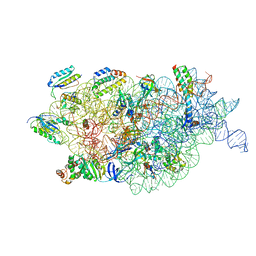 | | Bacterial 30S ribosomal subunit assembly complex state F (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
8V1F
 
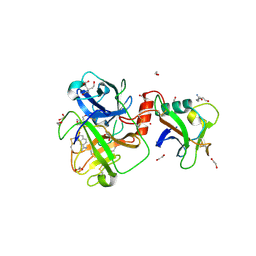 | | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Halabelian, L, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol
To Be Published
|
|
7B9H
 
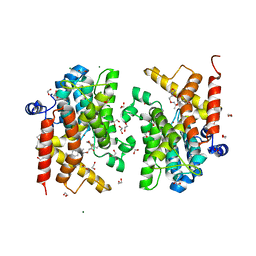 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-12-14 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
8U8O
 
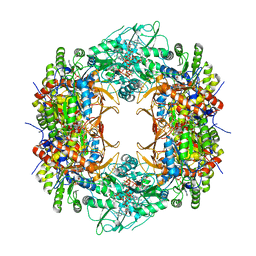 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
6U0P
 
 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
