6G6V
 
 | |
5Q1L
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000073a | | Descriptor: | (6~{R})-6-(4-methoxyphenyl)-2-oxidanylidene-5,6-dihydro-1~{H}-pyrimidine-4-carboxylic acid, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1Q8D
 
 | | The crystal structure of GDNF family co-receptor alpha 1 domain 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GDNF family receptor alpha 1 | | Authors: | Leppanen, V.M, Bespalov, M.M, Runeberg-Roos, P, Puurand, U, Merits, A, Saarma, M, Goldman, A. | | Deposit date: | 2003-08-21 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of GFRalpha1 domain 3 reveals new insights into GDNF binding and RET activation.
Embo J., 23, 2004
|
|
5Q1Q
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000294a | | Descriptor: | 8-[(dimethylamino)methyl]-4-methyl-7-oxidanyl-chromen-2-one, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3LRS
 
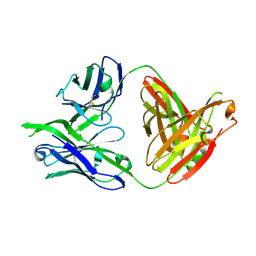 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
2X3F
 
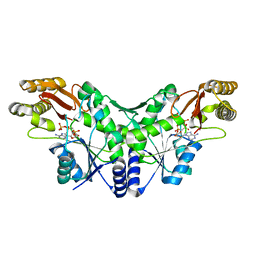 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
5Q1U
 
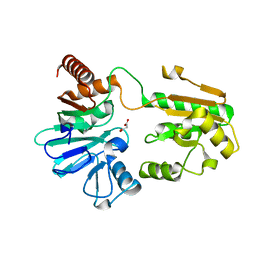 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000351a | | Descriptor: | DNA cross-link repair 1A protein, MALONATE ION, NICKEL (II) ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1AU9
 
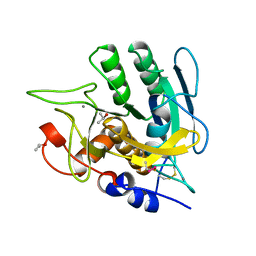 | | SUBTILISIN BPN' MUTANT 8324 IN CITRATE | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN BPN', ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-09-12 | | Release date: | 1997-12-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1QWD
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
1W9Y
 
 | | The structure of ACC oxidase | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE 1, SULFATE ION | | Authors: | Zhang, Z, Ren, J.-S, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2004-10-20 | | Release date: | 2005-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanistic Implications of 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (the Ethyling Forming Enzyme)
Chem.Biol., 11, 2004
|
|
1J7N
 
 | | Anthrax Toxin Lethal factor | | Descriptor: | Lethal Factor precursor, SULFATE ION, ZINC ION | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-05-17 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
2C3C
 
 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
5Q1P
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000299a | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5M4T
 
 | |
6EZL
 
 | |
6EXO
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
2BD0
 
 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
6EX8
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-~{N}-[1-(aminomethyl)cyclopropyl]-19-chloranyl-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
4YRK
 
 | |
1KSU
 
 | | Crystal Structure of His505Tyr Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
4YRP
 
 | |
1S6D
 
 | | Structure in solution of a methionine-rich 2S Albumin protein from Sunflower Seed | | Descriptor: | Albumin 8 | | Authors: | Pantoja-Uceda, D, Bruix, M, Shewry, P.R, Santoro, J, Rico, M. | | Deposit date: | 2004-01-23 | | Release date: | 2004-06-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Methionine-Rich 2S Albumin from Sunflower Seeds: Relationship to Its Allergenic and Emulsifying Properties.
Biochemistry, 43, 2004
|
|
1JKY
 
 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
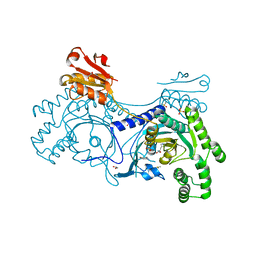
4YRO
 
 | |
