8QLF
 
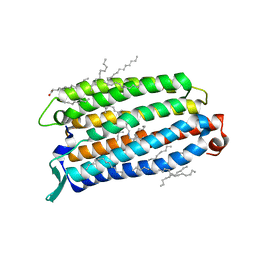 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QQZ
 
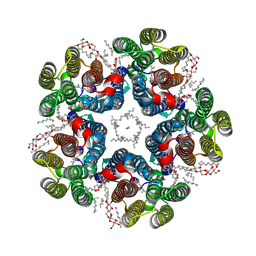 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
6ZLJ
 
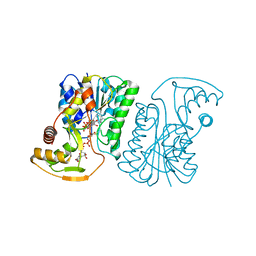 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase Y149F mutant from Bacillus cereus in complex with UDP-4-DEOXY-4-FLUORO-Glucuronic acid and NAD | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidany l)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-fluoranyl-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
4QGL
 
 | | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions | | Descriptor: | Acireductone dioxygenase, CADMIUM ION | | Authors: | Milaczewska, A.M, Chruszcz, M, Majorek, K.A, Porebski, P.J, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Acireductone dioxygenase from Bacillus anthracis with three cadmium ions
To be Published
|
|
8REV
 
 | |
6YXX
 
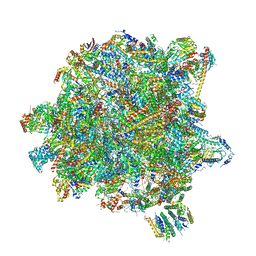 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | Descriptor: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | Authors: | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2020-05-04 | | Release date: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
5D0B
 
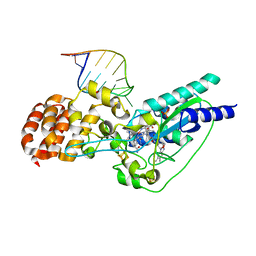 | | Crystal structure of epoxyqueuosine reductase with a tRNA-TYR epoxyqueuosine-modified tRNA stem loop | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, GLYCEROL, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5CX1
 
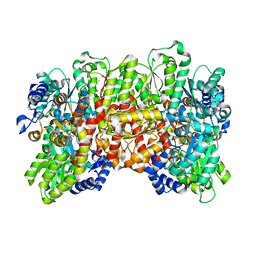 | | Nitrogenase molybdenum-iron protein beta-K400E mutant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Luca, M.A, Tezcan, F.A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7476 Å) | | Cite: | Evidence for Functionally Relevant Encounter Complexes in Nitrogenase Catalysis.
J.Am.Chem.Soc., 137, 2015
|
|
6ZL6
 
 | |
8QLE
 
 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
6ZLD
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid and NAD | | Descriptor: | Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
6ZLL
 
 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Savino, S, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
4L0E
 
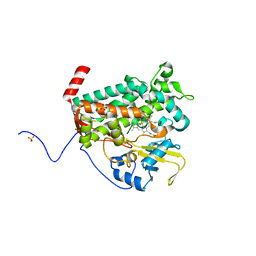 | |
6YC2
 
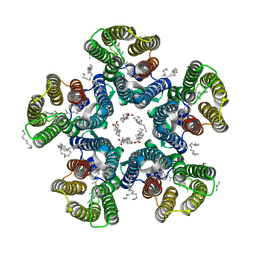 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form at room temperature, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ALANINE, EICOSANE, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
6ZLA
 
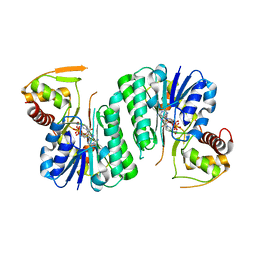 | |
6ZLK
 
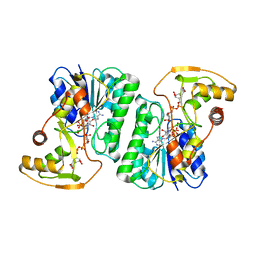 | | Equilibrium Structure of UDP-Glucuronic acid 4-epimerase from Bacillus cereus in complex with UDP-Glucuronic acid/UDP-Galacturonic acid and NAD | | Descriptor: | (2S,3R,4S,5R,6R)-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4,5-trihydroxy-oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
4L0F
 
 | |
8SPP
 
 | |
4P2B
 
 | |
5DQN
 
 | | Polyethylene 600-bound form of P450 CYP125A3 mutant from Myobacterium Smegmatis - W83Y | | Descriptor: | CITRIC ACID, Cytochrome P450 CYP125, PENTAETHYLENE GLYCOL, ... | | Authors: | Ortiz de Montellano, P.J, Frank, D.J, Waddling, C.A. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis.
Biochemistry, 54, 2015
|
|
4QGN
 
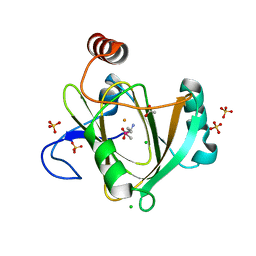 | | Human acireductone dioxygenase with iron ion and L-methionine in active center | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Milaczewska, A.M, Chruszcz, M, Petkowski, J.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2014-05-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | On the Structure and Reaction Mechanism of Human Acireductone Dioxygenase.
Chemistry, 24, 2018
|
|
4RRU
 
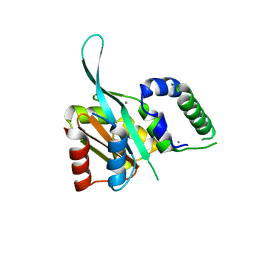 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
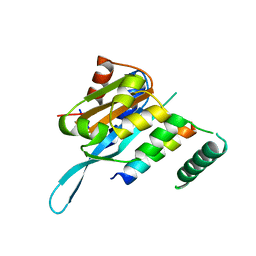 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
