6RU1
 
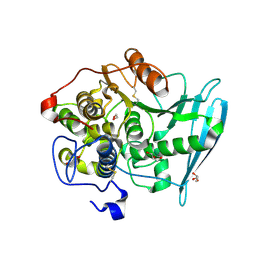 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
7Y60
 
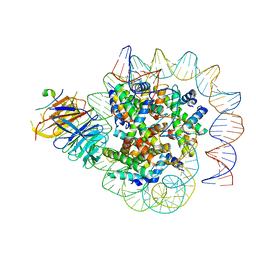 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8UTJ
 
 | | E. coli 70S ribosome with unmodified lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the 0 frame | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-31 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
4HT2
 
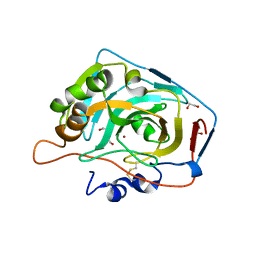 | | Crystal structure of human carbonic anhydrase isozyme XII with the inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4,6-dimethylpyrimidin-2-yl)thio]-2,3,5,6-tetrafluorobenzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 4-Substituted-2,3,5,6-tetrafluorobenzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, XII, and XIII.
Bioorg.Med.Chem., 21, 2013
|
|
8URM
 
 | | E. coli 70S ribosome with unmodified tRNAPro(GGG) bound to slippery P-site CCC-C codon and tRNAVal(UAC) in the A site | | Descriptor: | 13S ribosomal RNA, 16S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-10-26 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
8UXB
 
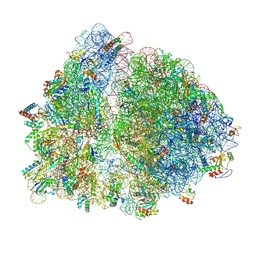 | | E. coli 70S ribosome with unmodified P/E-tRNAPro(GGG) bound to slippery P-site CCC-C codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
7G05
 
 | | Crystal Structure of human FABP4 in complex with 2,6-dichloro-4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenol, i.e. SMILES C(C(F)(F)F)(C(F)(F)F)(c1cc(c(c(c1)Cl)O)Cl)O with IC50=12 microM | | Descriptor: | 2,6-dichloro-4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenol, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FZ2
 
 | | Crystal Structure of human FABP4 in complex with [3-oxo-5-[4-(trifluoromethyl)phenyl]cyclohexen-1-yl] acetate, i.e. SMILES [C@H]1(c2ccc(cc2)C(F)(F)F)CC(=CC(=O)C1)OC(=O)C with IC50=0.833 microM | | Descriptor: | (1S)-5-hydroxy-4'-(trifluoromethyl)-1,6-dihydro[1,1'-biphenyl]-3(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kumagai, Y, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8CCS
 
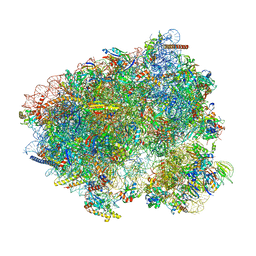 | | 80S S. cerevisiae ribosome with ligands in hybrid-1 pre-translocation (PRE-H1) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-27 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
1LN8
 
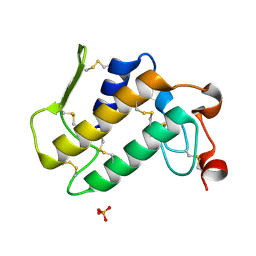 | | Crystal Structure of a New Isoform of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Phospholipase A2 | | Authors: | Singh, R.K, Vikram, P, Paramasivam, M, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-05-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a New Form of Phospholipase A2 from Naja naja sagittifera at 1.6 A Resolution
to be published
|
|
6HVS
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 18 | | Descriptor: | (2~{S})-~{N}-[2-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
5UUV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with a product IMP and the inhibitor P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with a product IMP and the inhibitor P182
To Be Published
|
|
8BQ6
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
3AQC
 
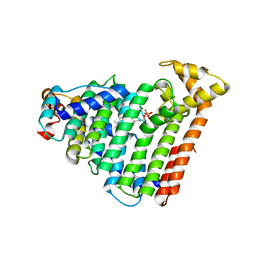 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium and FPP analogue | | Descriptor: | (2E,6E)-7,11-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
8WEA
 
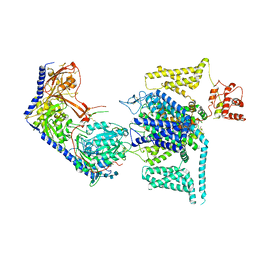 | | Human L-type voltage-gated calcium channel Cav1.2 (Class II) in the presence of pinaverium at 3.2 Angstrom resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
4HB9
 
 | |
1LF7
 
 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
4P5Z
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4LC8
 
 | | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4PTL
 
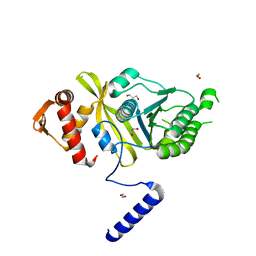 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-GM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4GRB
 
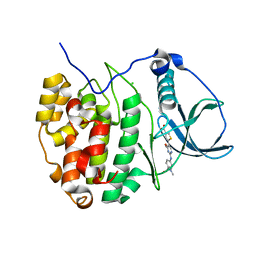 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
4ZLI
 
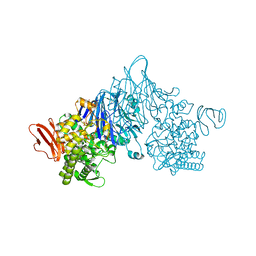 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4Z3X
 
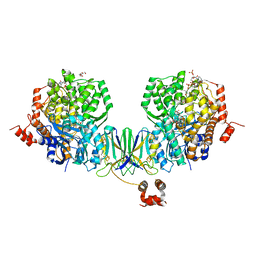 | | Active site complex BamBC of Benzoyl Coenzyme A reductase in complex with 1-Monoenoyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, Benzoyl-CoA reductase, putative, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
7JRA
 
 | |
5W2I
 
 | | Crystal structure of the core catalytic domain of human inositol phosphate multikinase soaked with C4-analogue of PtdIns(4,5)P2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol polyphosphate multikinase,Inositol polyphosphate multikinase, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2017-06-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities.
J. Biol. Chem., 292, 2017
|
|
