5NHB
 
 | |
6TJD
 
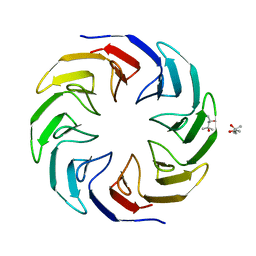 | | Crystal structure of the computationally designed Cake4 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake4 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
5NHN
 
 | |
6TJJ
 
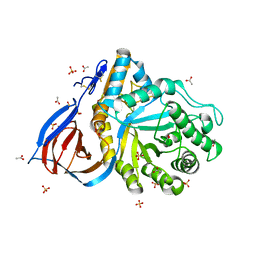 | | Structure of Cerezyme at pH 4.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6T99
 
 | |
6TKI
 
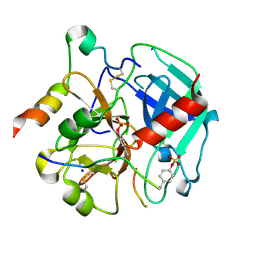 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - tetragonal form at 12.7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
5NHZ
 
 | | VIM-2_10b. Metallo-beta-Lactamase Inhibitors by Bioisosteric Replacement: Preparation, Activity and Binding | | Descriptor: | Beta-lactamase class B VIM-2, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Skagseth, S, Akhter, S, Paulsen, M.H, Samuelsen, O, Muhammad, Z, Leiros, K.-K.S, Bayer, A. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metallo-beta-lactamase inhibitors by bioisosteric replacement: Preparation, activity and binding.
Eur J Med Chem, 135, 2017
|
|
6T9Q
 
 | |
5N56
 
 | |
6TA2
 
 | | Human NAMPT in complex with nicotinic acid mononucleotide and phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
6TAD
 
 | | Bd0314 DslA E143Q mutant | | Descriptor: | SLT domain-containing protein | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus.
Nat Commun, 11, 2020
|
|
5N84
 
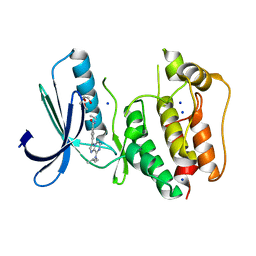 | | TTK kinase domain in complex with Mps-BAY2b | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dual specificity protein kinase TTK, SODIUM ION, ... | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-22 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5N8R
 
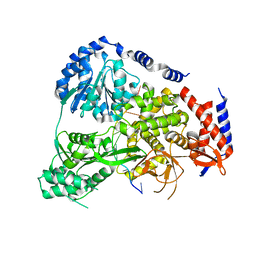 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | Descriptor: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9F
 
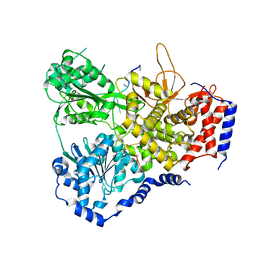 | | Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A | | Descriptor: | CG9323, isoform A, CHLORIDE ION, ... | | Authors: | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | Deposit date: | 2017-02-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.969 Å) | | Cite: | Structural and mechanistic insights into DHX36-mediated innate immunity and G-quadruplex unfolding
To Be Published
|
|
6SKT
 
 | |
5NA5
 
 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) apo structure | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-02-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and mechanistic basis of differentiated inhibitors of the acute pancreatitis target kynurenine-3-monooxygenase.
Nat Commun, 8, 2017
|
|
6SLW
 
 | | Fragment AZ-004 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, WW domain-containing transcription regulator protein 1 | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Leysen, S, Genet, S, Somsen, B, Patel, J, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SM7
 
 | | Crystal structure of SLA Reductase YihU from E. Coli | | Descriptor: | 3-sulfolactaldehyde reductase, BORIC ACID | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
Acs Catalysis, 2020
|
|
5NJA
 
 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
6SNQ
 
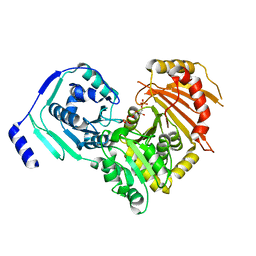 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Phosphoglucomutase-1, ZINC ION | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
5NK0
 
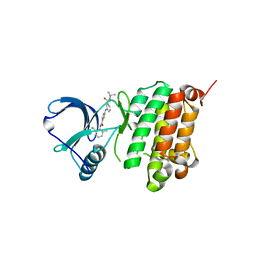 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1j | | Descriptor: | 2-[[3-[(3-azanyl-2,2-dimethyl-propyl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NK4
 
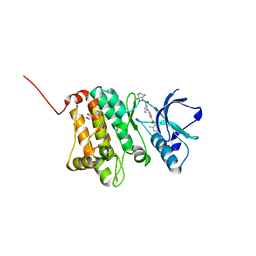 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2c | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-[[(4~{R})-3,3-bis(fluoranyl)piperidin-4-yl]carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6SPI
 
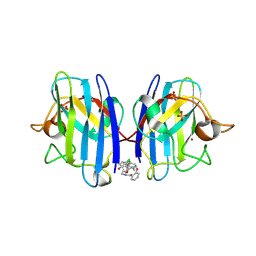 | | A4V MUTANT OF HUMAN SOD1 WITH EBSELEN DERIVATIVE 4 | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Chantadul, V, Amporndanai, K, Wright, G, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-09-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
5NKB
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4a | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3,5-dimorpholin-4-ylphenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKH
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3e | | Descriptor: | 2-[[3-(2-aminophenyl)-5-(piperidin-4-ylcarbamoyl)phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
