4LP8
 
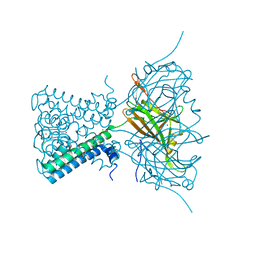 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
1LE5
 
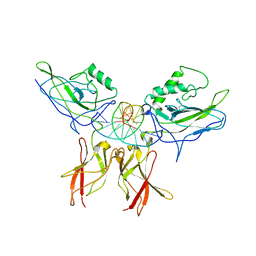 | | Crystal structure of a NF-kB heterodimer bound to an IFNb-kB | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*T)-3', Nuclear factor NF-kappa-B p50 subunit, ... | | Authors: | Berkowitz, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The X-ray crystal structure of the
NF-kB p50/p65 heterodimer bound
to the Interferon beta-kB site
J.Biol.Chem., 277, 2002
|
|
2GBT
 
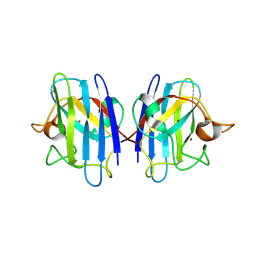 | | C6A/C111A CuZn Superoxide dismutase | | Descriptor: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
2GBU
 
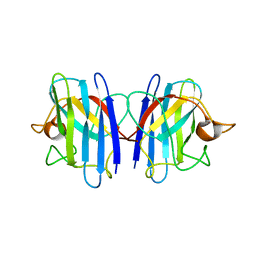 | | C6A/C111A/C57A/C146A apo CuZn Superoxide dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Hornberg, A, Logan, D.T, Marklund, S.L, Oliveberg, M. | | Deposit date: | 2006-03-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Coupling between Disulphide Status, Metallation and Dimer Interface Strength in Cu/Zn Superoxide Dismutase
J.Mol.Biol., 365, 2007
|
|
1PL4
 
 | | Crystal Structure of human MnSOD Y166F mutant | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
4HWW
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 9 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
2MF2
 
 | |
3KK6
 
 | | Crystal Structure of Cyclooxygenase-1 in complex with celecoxib | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, CITRATE ANION, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2009-11-04 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4MPR
 
 | |
5MG8
 
 | | Crystal structure of the S.pombe Smc5/6 hinge domain | | Descriptor: | GLYCEROL, SULFATE ION, Structural maintenance of chromosomes protein 5, ... | | Authors: | Alt, A, Pearl, L.H, Oliver, A.W. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Specialized interfaces of Smc5/6 control hinge stability and DNA association.
Nat Commun, 8, 2017
|
|
5MAL
 
 | |
2KIV
 
 | | AIDA-1 SAM domain tandem | | Descriptor: | Ankyrin repeat and sterile alpha motif domain-containing protein 1B | | Authors: | Donaldson, L.W, Kurabi, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A nuclear localization signal at the SAM-SAM domain interface of AIDA-1 suggests a requirement for domain uncoupling prior to nuclear import.
J.Mol.Biol., 392, 2009
|
|
4HXQ
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 14 | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-carboxy-5-(methylamino)-7-(piperidin-1-yl)heptyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
4O41
 
 | | Amide linked RNA | | Descriptor: | AMIDE LINKED RNA, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs.
Nucleic Acids Res., 42, 2014
|
|
2R3B
 
 | |
2R3E
 
 | |
5NFQ
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5VAJ
 
 | | BhRNase H - amide-RNA/DNA complex | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
4FDZ
 
 | | EutL from Clostridium perfringens, Crystallized Under Reducing Conditions | | Descriptor: | Ethanolamine utilization protein, SODIUM ION | | Authors: | Thompson, M.C, Cascio, D, Crowley, C.S, Kopstein, J.S, Yeates, T.O. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein.
Protein Sci., 24, 2015
|
|
1EL6
 
 | | STRUCTURE OF BACTERIOPHAGE T4 GENE PRODUCT 11, THE INTERFACE BETWEEN THE BASEPLATE AND SHORT TAIL FIBERS | | Descriptor: | BASEPLATE STRUCTURAL PROTEIN GP11 | | Authors: | Leiman, P.G, Kostyuchenko, V.A, Schneider, M.M, Kurochkina, L.P, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2000-03-13 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of bacteriophage T4 gene product 11, the interface between the baseplate and short tail fibers.
J.Mol.Biol., 301, 2000
|
|
3FVI
 
 | | Crystal Structure of Complex of Phospholipase A2 with Octyl Sulfates | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Pan, Y.H. | | Deposit date: | 2009-01-15 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a premicellar complex of alkyl sulfates with the interfacial binding surfaces of four subunits of phospholipase A2.
Biochim.Biophys.Acta, 1804, 2010
|
|
2LUV
 
 | |
5NRF
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7i | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-chlorophenyl)ethyl]-~{N}-(phenylmethyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
1LE9
 
 | | Crystal structure of a NF-kB heterodimer bound to the Ig/HIV-kB siti | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*T)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Benjamin, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The x-ray crystal structure of the NF-kappa B p50.p65 heterodimer bound to the interferon beta -kappa B site.
J.Biol.Chem., 277, 2002
|
|
3PHY
 
 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Dux, P, Rubinstenn, G, Vuister, G.W, Boelens, R, Mulder, F.A.A, Hard, K, Hoff, W.D, Kroon, A, Crielaard, W, Hellingwerf, K.J, Kaptein, R. | | Deposit date: | 1998-02-06 | | Release date: | 1998-05-27 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the photoactive yellow protein.
Biochemistry, 37, 1998
|
|
