2B1A
 
 | | Crystal structure analysis of anti-HIV-1 V3 Fab 2219 in complex with UG1033 peptide | | Descriptor: | Fab 2219, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity.
J.Virol., 80, 2006
|
|
5VOZ
 
 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 3) | | Descriptor: | Uncharacterized protein, V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
2PMU
 
 | | Crystal structure of the DNA-binding domain of PhoP | | Descriptor: | CHLORIDE ION, GLYCINE, PHOSPHATE ION, ... | | Authors: | Wang, S. | | Deposit date: | 2007-04-23 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator PhoP from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2PHH
 
 | |
5BX3
 
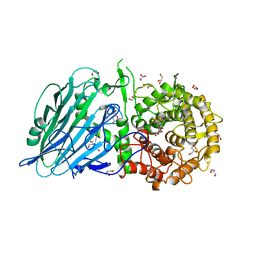 | | Crystal structure of Thermoanaerobacterium xylanolyticum GH116 beta-glucosidase with deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, CALCIUM ION, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tankrathok, A, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-08 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2)
Acs Chem.Biol., 11, 2016
|
|
6TTT
 
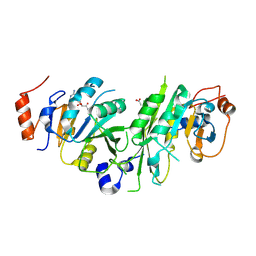 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 2 (ASI_M3M_140) | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-~{N}-methyl-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
2G32
 
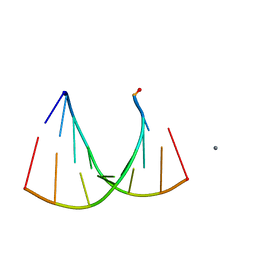 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2PHN
 
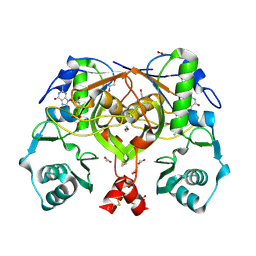 | | Crystal structure of an amide bond forming F420-gamma glutamyl ligase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, F420-0:gamma-glutamyl ligase, ... | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an Amide Bond Forming F(420):gammagamma-glutamyl Ligase from Archaeoglobus Fulgidus - A Member of a New Family of Non-ribosomal Peptide Synthases.
J.Mol.Biol., 372, 2007
|
|
6TOW
 
 | |
4EVI
 
 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol 9-Methyl Ether and S -Adenosyl-L-Homocysteine | | Descriptor: | 2-methoxy-4-[(1E)-3-methoxyprop-1-en-1-yl]phenol, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, ... | | Authors: | Wolters, S, Heine, A, Petersen, M. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2PLA
 
 | | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein | | Descriptor: | CHLORIDE ION, Glycerol-3-phosphate dehydrogenase 1-like protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Uppenberg, J, Smee, C, Hozjan, V, Kavanagh, K, Bunkoczi, G, Papagrigoriou, E, Pike, A.C.W, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of human glycerol-3-phosphate dehydrogenase 1-like protein.
To be Published
|
|
5W2Q
 
 | | Crystal structure of Mycobacterium tuberculosis KasA in complex with 6U5 | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 1, GLYCEROL, ... | | Authors: | Capodagli, G.C, Neiditch, M.B. | | Deposit date: | 2017-06-06 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synergistic Lethality of a Binary Inhibitor of Mycobacterium tuberculosis KasA.
MBio, 9, 2018
|
|
2FZB
 
 | | Human Aldose Reductase complexed with four tolrestat molecules at 1.5 A resolution. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
5BVU
 
 | | Crystal structure of Thermoanaerobacterium xylolyticum GH116 beta-glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Charoenwattanasatien, R, Pengthaisong, S, Sansenya, S, Mutoh, R, Tanaka, H, Kurisu, G, Ketudat Cairns, J.R. | | Deposit date: | 2015-06-05 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bacterial beta-Glucosidase Reveals the Structural and Functional Basis of Genetic Defects in Human Glucocerebrosidase 2 (GBA2).
Acs Chem.Biol., 11, 2016
|
|
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
4ZUK
 
 | | Structure ALDH7A1 complexed with NAD+ | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
2PS7
 
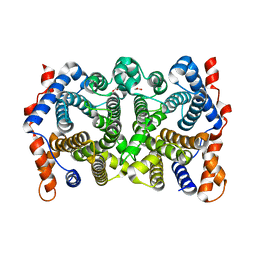 | | Y295F trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Trichodiene synthase | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-05-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif.
Arch.Biochem.Biophys., 469, 2008
|
|
2PSN
 
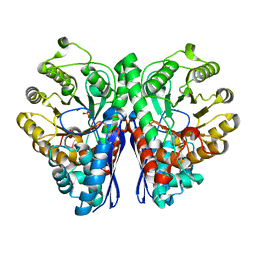 | |
4ZW3
 
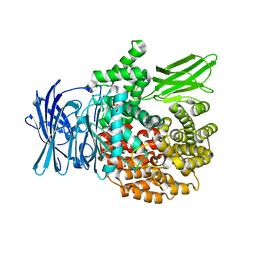 | |
2PTW
 
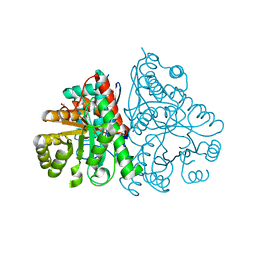 | | Crystal Structure of the T. brucei enolase complexed with sulphate, identification of a metal binding site IV | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
2YGA
 
 | | E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GELDANAMYCIN | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Features of the Streptomyces Hygroscopicus Htpg Reveal How Partial Geldanamycin Resistance Can Arise with Mutation to the ATP Binding Pocket of a Eukaryotic Hsp90.
Faseb J., 25, 2011
|
|
4ZXV
 
 | |
6TRH
 
 | |
4ZX4
 
 | |
2JZT
 
 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
